Prediction of the Outcome for Patients with Glioblastoma with lncRNA Expression Profiles - PubMed (original) (raw)
Prediction of the Outcome for Patients with Glioblastoma with lncRNA Expression Profiles
Qinglin Liu et al. Biomed Res Int. 2019.
Abstract
Background: Progress in gene sequencing has paved the way for precise outcome prediction of the heterogeneous disease of glioblastoma. The aim was to assess the potential of utilizing the lncRNA expression profile for predicting glioblastoma patient survival.
Materials and methods: Clinical and lncRNA expression data were downloaded from the public database of the cancer genome atlas. Differentially expressed lncRNAs between glioblastoma and normal brain tissue were screened by bioinformatics analysis. The samples were randomly separated into the training and testing sets. Univariate Cox regression, least absolute shrinkage, selection operator regression, and multivariate Cox regression were performed to develop the prediction model with the training set, which was presented as a forest plot. The performance of the model was validated by discrimination and calibration analysis in both the training and testing sets. Patient survival between model-predicted low- and high-risk subgroups was compared in both the training and testing sets.
Results: One thousand two hundred and fifty-five differentially expressed lncRNAs between glioblastoma and normal brain tissues were screened. After univariate Cox regression and the least absolute shrinkage and selection operator regression, a 12 lncRNA constituted prediction model was developed by multivariate Cox regression. Of the 12 lncRNAs, 4 lncRNAs were independent risk factors for patient survival. The areas under the receiver operating characteristic curves of the model for predicting 0.5-, 1-, 1.5-, and 2-year patient survival was 0.788, 0.824, 0.874, and 0.886, respectively in the training set and 0.723, 0.84, 0.816, and 0.773 in the testing set. The calibration curves of the prediction model fitted well. Significant survival disparity was observed between the model dichotomized low- and high-risk subgroups in both the training and testing set.
Conclusions: LncRNA expression signature can predict glioblastoma patient survival, promising lncRNA-based survival prediction.
Copyright © 2019 Qinglin Liu et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures
Figure 1
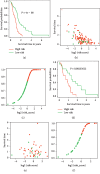
Heatmap showed the top 50 differentially expressed lncRNAs between GBM and normal brain tissue.
Figure 2
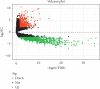
Volcano plot showed the differentially expressed lncRNAs between GBM and normal brain tissues. Black dots: not differentially expressed lncRNAs, Green dots: downregulated lncRNAs with expression level P < 0.05 and fold of change >2 and red dots: upregulated lncRNAs with expression level P < 0.05 and fold of change >2.
Figure 3
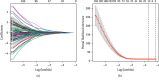
Feature selection using the least absolute shrinkage and selection operator (LASSO) binary logistic regression model. LASSO coefficient profiles of the 132 differentially expressed lncRNAs (a). The tuning parameter (λ) selection in the LASSO model used 10-fold cross-validation via minimum criteria (b). A coefficient profile plot was produced against the log (λ) sequence. The vertical lines were drawn at the value of the minimum and minimum + 1 standard error selected using 10-fold cross-validation, where optimal λ resulted in 12 nonzero coefficients.
Figure 4
The constitution of the prediction model demonstrated as a forest plot. Four lncRNAs were independent risk factors for patient survival.
Figure 5
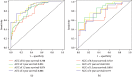
Performance of the prediction model. Receiver-operating characteristic curves (ROCs) for the model in predicting the 0.5-, 1-, 1.5-, and 2-year survival were built, and the prediction accuracy was demonstrated with areas under the curves (AUCs) in the training (a) and testing set (b).
Figure 6
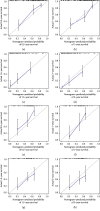
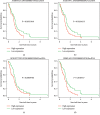
Calibration plots of the model for predicting 0.5- (a), 1- (b), 1.5- (c), and 2-year (d) survival in the training set. Calibration plots of the model for predicting 0.5- (e), 1- (f), 1.5- (g), and 2-year (h) survival in the testing set.
Figure 7
Risk score analysis of the patients. Survival analysis of the GBM patients dichotomized by the model-predicted risk scores in the training (a) long-rank test (P < 0.01) and testing set ((d) long-rank test, _P_=0.002). The patient's status along with model-predicted risk scores in the training (b) and testing set (e). Model-predicted risk score distribution of GBM patients in the training (c) and testing (f) set.
Figure 8
Survival analysis of GBM patients dichotomized by independent lncRNAs associated with overall survival in the training set. Survival analysis of the GBM patients dichotomized by AC005632.4.ENSG00000273956.lincRNA (a). Survival analysis of the GBM patients dichotomized by AC021594.1.ENSG00000266924.lincRNA (b). Survival analysis of the GBM patients dichotomized by MIRLET7DHG.ENSG00000230262.lincRNA (c). Survival analysis of the GBM patients dichotomized by OSMR.AS1.ENSG00000249740.lincRNA (d).
Similar articles
- A novel seven-lncRNA signature for prognosis prediction in hepatocellular carcinoma.
Yan J, Zhou C, Guo K, Li Q, Wang Z. Yan J, et al. J Cell Biochem. 2019 Jan;120(1):213-223. doi: 10.1002/jcb.27321. Epub 2018 Sep 11. J Cell Biochem. 2019. PMID: 30206981 - Discovery of a novel six-long non-coding RNA signature predicting survival of colorectal cancer patients.
Fan Q, Liu B. Fan Q, et al. J Cell Biochem. 2018 Apr;119(4):3574-3585. doi: 10.1002/jcb.26548. Epub 2018 Jan 15. J Cell Biochem. 2018. PMID: 29227531 - A Novel Immune-Related lncRNA-Based Model for Survival Prediction in Clear Cell Renal Cell Carcinoma.
Zhang Z, Tang Y, Liu Y, Zhuang H, Lin E, Xie L, Feng X, Tian K, Zeng J, Liu J, Yu Y. Zhang Z, et al. J Immunol Res. 2021 Jun 28;2021:9921466. doi: 10.1155/2021/9921466. eCollection 2021. J Immunol Res. 2021. PMID: 34368371 Free PMC article. - An eight-mRNA signature outperforms the lncRNA-based signature in predicting prognosis of patients with glioblastoma.
Gong Z, Hong F, Wang H, Zhang X, Chen J. Gong Z, et al. BMC Med Genet. 2020 Mar 19;21(1):56. doi: 10.1186/s12881-020-0992-7. BMC Med Genet. 2020. PMID: 32188434 Free PMC article. - Post-Transcriptional Modifications of RNA as Regulators of Apoptosis in Glioblastoma.
Dome A, Dymova M, Richter V, Stepanov G. Dome A, et al. Int J Mol Sci. 2022 Aug 17;23(16):9272. doi: 10.3390/ijms23169272. Int J Mol Sci. 2022. PMID: 36012529 Free PMC article. Review.
Cited by
- Large-Scale Analysis Reveals Gene Signature for Survival Prediction in Primary Glioblastoma.
Prasad B, Tian Y, Li X. Prasad B, et al. Mol Neurobiol. 2020 Dec;57(12):5235-5246. doi: 10.1007/s12035-020-02088-w. Epub 2020 Sep 1. Mol Neurobiol. 2020. PMID: 32875483 Free PMC article. - A Prognostic Model for Brain Glioma Patients Based on 9 Signature Glycolytic Genes.
Bingxiang X, Panxing W, Lu F, Xiuyou Y, Chao D. Bingxiang X, et al. Biomed Res Int. 2021 Jun 16;2021:6680066. doi: 10.1155/2021/6680066. eCollection 2021. Biomed Res Int. 2021. PMID: 34222480 Free PMC article. Retracted. - An in silico glioblastoma microenvironment model dissects the immunological mechanisms of resistance to PD-1 checkpoint blockade immunotherapy.
Zhang Z, Liu L, Ma C, Cui X, Lam RHW, Chen W. Zhang Z, et al. Small Methods. 2021 Jun 15;5(6):2100197. doi: 10.1002/smtd.202100197. Epub 2021 Apr 22. Small Methods. 2021. PMID: 34423116 Free PMC article. - Long Non-Coding RNA UBA6-AS1 Promotes the Malignant Properties of Glioblastoma by Competitively Binding to microRNA-760 and Enhancing Homeobox A2 Expression.
Cheng F, Liu J, Zhang Y, You Q, Chen B, Cheng J, Deng C. Cheng F, et al. Cancer Manag Res. 2021 Jan 14;13:379-392. doi: 10.2147/CMAR.S287676. eCollection 2021. Cancer Manag Res. 2021. PMID: 33469379 Free PMC article. Retracted.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources