Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles - PubMed (original) (raw)
Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles
Gints Kalnins et al. Nat Commun. 2020.
Abstract
Bacterial microcompartments (BMCs) are prokaryotic organelles consisting of a protein shell and an encapsulated enzymatic core. BMCs are involved in several biochemical processes, such as choline, glycerol and ethanolamine degradation and carbon fixation. Since non-native enzymes can also be encapsulated in BMCs, an improved understanding of BMC shell assembly and encapsulation processes could be useful for synthetic biology applications. Here we report the isolation and recombinant expression of BMC structural genes from the Klebsiella pneumoniae GRM2 locus, the investigation of mechanisms behind encapsulation of the core enzymes, and the characterization of shell particles by cryo-EM. We conclude that the enzymatic core is encapsulated in a hierarchical manner and that the CutC choline lyase may play a secondary role as an adaptor protein. We also present a cryo-EM structure of a pT = 4 quasi-symmetric icosahedral shell particle at 3.3 Å resolution, and demonstrate variability among the minor shell forms.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1. Klebsiella pneumoniae GRM2 locus and variants of cmcC.
a Klebsiella pneumonia GRM2 locus. Structural shell BMC-H proteins cmcA, cmcB, cmcC, and cmcE are colored in green, and BMC-P protein cmcD is colored in yellow. Core enzymes CutF (aldehyde dehydrogenase), CutO (alcohol dehydrogenase), CutC (choline lyase), CutD (glycyl-radical activating enzyme), and CutH (phosphotransacylase) are colored in blue. Regulatory and transporter genes are colored in gray. The genes have been named according to previous research. b C-terminal amino acid sequences of three cmcC variants—cmcC (native), cmcC′ (mutated), and cmcCtrunc (truncated).
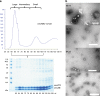
Fig. 2. Characterization of cmcABC′ + cmcD BDPs.
a Gel filtration of sedimented BDPs and SDS-PAGE analysis of the fractions. Two noticeable BDP peaks were formed: one immediately after the empty volume of 60 ml (large type particle zone) and one at approximately 90–100 ml (small type particle zone). An intermediary zone with smaller BDP protein amounts was formed between these two zones. b Examples of TEM analysis of the BDP samples in the large particle zone (66 ml), intermediary zone (76 and 84 ml), and small particle zone (96 ml). Scale bar: 200 nm.
Fig. 3. Proposed enzymatic core encapsulation mechanism of CutC, CutF, and CutO in GRM2 BDPs.
CutC is serving as an adaptor for the encapsulation of other enzymes. CutC C-terminal domain is responsible for encapsulation and also for interaction with CutO. The CutC N-terminal domain is responsible for CutC interaction with CutF. CutF together with CutC N-terminal domain crosslinks the enzymatic core and increases its size.
Fig. 4. Cryo-EM structure of pT=4 quasi-icosahedral BDP and its penatameric and hexameric components.
a Surface model of pT = 4 quasi-icosahedral BDP particle, displayed on the left side. A ribbon model of a cmcD pentamer and three cmcC′ hexamers is displayed on the right side. Pentameric cmcD protein is colored in yellow and hexameric cmcC′ is colored in green. Note that the fivefold symmetry axis is located at the center of cmcD pentamer and threefold axis is located in the middle between three cmcC′ hexamers. b Electrostatic surface potential of pentameric cmcD and hexameric cmcC′. Note the pores in the centers of pentamers and hexamers. The surface contour levels were set to −1 kT/e (red) and +1 kT/e (blue).
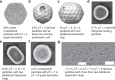
Fig. 5. Cryo-EM classes of BDP subtypes.
a 3D class at 3.3 Å resolution and atomic model of whole intact pT = 4 quasi-symmetric icosahedral particles, in total 69%. b 8.8 Å resolution 3D class of pT = 4 derived particles, with at least one missing pentameric unit, in total 23%. c 9.6 Å resolution 3D class of pT = 4, Q = 6 quasi-symmetric particles, in total 6%. d 2D class of triangular-looking particles, judging from the size probably derived from three fused pT = 4 particles, in total 0.7%. e 2D class of pT = 4, Q = 8 particles, in total 0.3%. f 2D class of pT = 7 or pT = 9 quasi-icosahedral particles, in total 0.2%. g Separate micrographs of tubular particles elongated with more than two hexameric rings, in total less than 0.1%. Scale bar: 60 nm.
Fig. 6. Detailed view of hexameric–hexameric and pentameric–hexameric interfaces.
a Hexameric–hexameric cmcC′–cmcC′ interface; K–R–X (in our case K–R–A) triad and salt bridge networks are viewed as stick models, distances are measured in angstroms. b Hexameric–pentameric cmcC′–cmcD interface; amino acids involved in contacts are viewed as stick models. Distances are measured in angstroms.
Similar articles
- Variety of size and form of GRM2 bacterial microcompartment particles.
Cesle EE, Filimonenko A, Tars K, Kalnins G. Cesle EE, et al. Protein Sci. 2021 May;30(5):1035-1043. doi: 10.1002/pro.4069. Epub 2021 Apr 2. Protein Sci. 2021. PMID: 33763934 Free PMC article. - Modulation of Hybrid GRM2-type Bacterial Microcompartment Shells through BMC-H Shell Protein Fusion and Incorporation of Non-native BMC-T Shell Proteins.
Česle EEL, Ta Rs K, Jansons J, Kalniņš G. Česle EEL, et al. ACS Synth Biol. 2023 Nov 17;12(11):3275-3286. doi: 10.1021/acssynbio.3c00281. Epub 2023 Nov 8. ACS Synth Biol. 2023. PMID: 37937366 - Effect of metabolosome encapsulation peptides on enzyme activity, coaggregation, incorporation, and bacterial microcompartment formation.
Juodeikis R, Lee MJ, Mayer M, Mantell J, Brown IR, Verkade P, Woolfson DN, Prentice MB, Frank S, Warren MJ. Juodeikis R, et al. Microbiologyopen. 2020 May;9(5):e1010. doi: 10.1002/mbo3.1010. Epub 2020 Feb 13. Microbiologyopen. 2020. PMID: 32053746 Free PMC article. - Protein stoichiometry, structural plasticity and regulation of bacterial microcompartments.
Liu LN, Yang M, Sun Y, Yang J. Liu LN, et al. Curr Opin Microbiol. 2021 Oct;63:133-141. doi: 10.1016/j.mib.2021.07.006. Epub 2021 Jul 31. Curr Opin Microbiol. 2021. PMID: 34340100 Review. - Dynamic structural determinants in bacterial microcompartment shells.
Trettel DS, Kerfeld CA, Gonzalez-Esquer CR. Trettel DS, et al. Curr Opin Microbiol. 2024 Aug;80:102497. doi: 10.1016/j.mib.2024.102497. Epub 2024 Jun 21. Curr Opin Microbiol. 2024. PMID: 38909546 Review.
Cited by
- Engineered bacterial microcompartments: apps for programming metabolism.
Kerfeld CA, Sutter M. Kerfeld CA, et al. Curr Opin Biotechnol. 2020 Oct;65:225-232. doi: 10.1016/j.copbio.2020.05.001. Epub 2020 Jun 15. Curr Opin Biotechnol. 2020. PMID: 32554213 Free PMC article. Review. - Heterologous Assembly of Pleomorphic Bacterial Microcompartment Shell Architectures Spanning the Nano- to Microscale.
Ferlez BH, Kirst H, Greber BJ, Nogales E, Sutter M, Kerfeld CA. Ferlez BH, et al. Adv Mater. 2023 Jun;35(23):e2212065. doi: 10.1002/adma.202212065. Epub 2023 Apr 25. Adv Mater. 2023. PMID: 36932732 Free PMC article. - Functionalization of bacterial microcompartment shell interior with cysteine containing peptides enhances the iron and cobalt loading capacity.
Kalnins G, Bertins M, Viksna A, Tars K. Kalnins G, et al. Biometals. 2024 Feb;37(1):267-274. doi: 10.1007/s10534-023-00538-1. Epub 2023 Sep 20. Biometals. 2024. PMID: 37728832 - Modeling bacterial microcompartment architectures for enhanced cyanobacterial carbon fixation.
Trettel DS, Pacheco SL, Laskie AK, Gonzalez-Esquer CR. Trettel DS, et al. Front Plant Sci. 2024 Feb 15;15:1346759. doi: 10.3389/fpls.2024.1346759. eCollection 2024. Front Plant Sci. 2024. PMID: 38425792 Free PMC article. Review. - Comparative Pore Structure and Dynamics for Bacterial Microcompartment Shell Protein Assemblies in Sheets or Shells.
Raza S, Sarkar D, Chan LJG, Mae J, Sutter M, Petzold CJ, Kerfeld CA, Ralston CY, Gupta S, Vermaas JV. Raza S, et al. bioRxiv [Preprint]. 2024 Mar 12:2024.03.12.584231. doi: 10.1101/2024.03.12.584231. bioRxiv. 2024. PMID: 38559214 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources