A pneumonia outbreak associated with a new coronavirus of probable bat origin - PubMed (original) (raw)
. 2020 Mar;579(7798):270-273.
doi: 10.1038/s41586-020-2012-7. Epub 2020 Feb 3.
Xing-Lou Yang # 1, Xian-Guang Wang # 2, Ben Hu 1, Lei Zhang 1, Wei Zhang 1, Hao-Rui Si 1 3, Yan Zhu 1, Bei Li 1, Chao-Lin Huang 2, Hui-Dong Chen 2, Jing Chen 1 3, Yun Luo 1 3, Hua Guo 1 3, Ren-Di Jiang 1 3, Mei-Qin Liu 1 3, Ying Chen 1 3, Xu-Rui Shen 1 3, Xi Wang 1 3, Xiao-Shuang Zheng 1 3, Kai Zhao 1 3, Quan-Jiao Chen 1, Fei Deng 1, Lin-Lin Liu 4, Bing Yan 1, Fa-Xian Zhan 4, Yan-Yi Wang 1, Geng-Fu Xiao 1, Zheng-Li Shi 5
Affiliations
- PMID: 32015507
- PMCID: PMC7095418
- DOI: 10.1038/s41586-020-2012-7
A pneumonia outbreak associated with a new coronavirus of probable bat origin
Peng Zhou et al. Nature. 2020 Mar.
Erratum in
- Addendum: A pneumonia outbreak associated with a new coronavirus of probable bat origin.
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. Zhou P, et al. Nature. 2020 Dec;588(7836):E6. doi: 10.1038/s41586-020-2951-z. Nature. 2020. PMID: 33199918 Free PMC article. No abstract available.
Abstract
Since the outbreak of severe acute respiratory syndrome (SARS) 18 years ago, a large number of SARS-related coronaviruses (SARSr-CoVs) have been discovered in their natural reservoir host, bats1-4. Previous studies have shown that some bat SARSr-CoVs have the potential to infect humans5-7. Here we report the identification and characterization of a new coronavirus (2019-nCoV), which caused an epidemic of acute respiratory syndrome in humans in Wuhan, China. The epidemic, which started on 12 December 2019, had caused 2,794 laboratory-confirmed infections including 80 deaths by 26 January 2020. Full-length genome sequences were obtained from five patients at an early stage of the outbreak. The sequences are almost identical and share 79.6% sequence identity to SARS-CoV. Furthermore, we show that 2019-nCoV is 96% identical at the whole-genome level to a bat coronavirus. Pairwise protein sequence analysis of seven conserved non-structural proteins domains show that this virus belongs to the species of SARSr-CoV. In addition, 2019-nCoV virus isolated from the bronchoalveolar lavage fluid of a critically ill patient could be neutralized by sera from several patients. Notably, we confirmed that 2019-nCoV uses the same cell entry receptor-angiotensin converting enzyme II (ACE2)-as SARS-CoV.
Conflict of interest statement
The authors declare no competing interests.
Figures
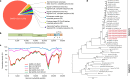
Fig. 1. Genome characterization of 2019-nCoV.
a, Metagenomics analysis of next-generation sequencing of BALF from patient ICU06. b, Genomic organization of 2019-nCoV WIV04. M, membrane. c, Similarity plot based on the full-length genome sequence of 2019-nCoV WIV04. Full-length genome sequences of SARS-CoV BJ01, bat SARSr-CoV WIV1, bat coronavirus RaTG13 and ZC45 were used as reference sequences. d, Phylogenetic tree based on nucleotide sequences of complete genomes of coronaviruses. MHV, murine hepatitis virus; PEDV, porcine epidemic diarrhoea virus; TGEV, porcine transmissible gastroenteritis virus.The scale bars represent 0.1 substitutions per nucleotide position. Descriptions of the settings and software that was used are included in the Methods.
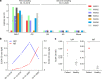
Fig. 2. Molecular and serological investigation of patient samples.
a, Molecular detection of 2019-nCoV in seven patients. Patient information can be found in Extended Data Tables 1, 2. Detection methods are described in the Methods. AS, anal swab; OS, oral swab. b, Dynamics of 2019-nCoV antibody levels in one patient who showed signs of disease on 23 December 2019 (ICU-06). OD ratio, optical density at 450–630 nm. The right and left y axes indicate ELISA OD ratios for IgM and IgG, respectively. c, Serological test of 2019-nCoV antibodies in five patients (Extended Data Table 2). The asterisk indicates data collected from patient ICU-06 on 10 January 2020. b, c, The cut-off was to 0.2 for the IgM analysis and to 0.3 for the IgG analysis, according to the levels of healthy controls.
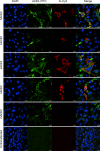
Fig. 3. Analysis of the receptor use of 2019-nCoV.
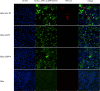
Determination of virus infectivity in HeLa cells that expressed or did not express (untransfected) ACE2. The expression of ACE2 plasmid with S tag was detected using mouse anti-S tag monoclonal antibody. hACE2, human ACE2; bACE2, ACE2 of Rhinolophus sinicus (bat); cACE2, civet ACE2; sACE2, swine ACE2 (pig); mACE2, mouse ACE2. Green, ACE2; red, viral protein (N); blue, DAPI (nuclei). Scale bars, 10 μm.
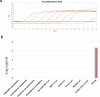
Extended Data Fig. 1. NGS raw reads of sample WIV04 mapping to the 2019-nCoV sequence.
The x axis indicates the genome nucleotide position and the y axis represents the read depth of the mapping.
Extended Data Fig. 2. Phylogenetic trees based on the complete S and RdRp gene sequences of coronaviruses.
a, b, Phylogenetic trees on the basis of the gene sequences of S (a) and RdRp (b) are shown. 2019-nCoV and bat CoV RaTG13 are shown in bold and in red. The trees were constructed using the maximum likelihood method using the GTR + G substitution model with bootstrap values determined by 1,000 replicates. Bootstraps values of more than 50% are shown.
Extended Data Fig. 3. Amino acid sequence alignment of the S1 protein of the 2019-nCoV to SARS-CoV and selected bat SARSr-CoVs.
The receptor-binding motif of SARS-CoV and the homologous region of other coronaviruses are indicated by the red box. The key amino acid residues involved in the interaction with human ACE2 are numbered at the top of the aligned sequences. The short insertions in the N-terminal domain of the 2019-nCoV are indicated by the blue boxes. Bat CoV RaTG13 was obtained from R. affinis, found in Yunnan province. Bat CoV ZC45 was obtained from R. sinicus, found in Zhejiang province.
Extended Data Fig. 4. Molecular detection method used to detect 2019-nCoV.
a, Standard curve for qPCR primers. The PCR product of the S gene that was serial diluted in the range of 108 to 101 (lines from left to right) was used as a template. Primer sequences and experimental conditions are described in the Methods. b, Specificity of the qPCR primers. Nucleotide samples from the indicated pathogens were used.
Extended Data Fig. 5. Amino acid sequence alignment of the nucleocapsid protein of 2019-nCoV to bat SARSr-CoV Rp3 and SARS-CoV BJ01.
Bat SARSr-CoV Rp3 was obtained from R. sinicus, which is found in Guangxi province.
Extended Data Fig. 6. Isolation and antigenic characterization of 2019-nCoV.
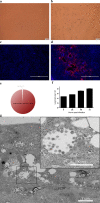
a, b, Vero E6 cells are shown at 24 h after infection with mock virus (a) or 2019-nCoV (b). c, d, Mock-virus-infected (c) or 2019-nCoV-infected (d) samples were stained with rabbit serum raised against recombinant SARSr-CoV Rp3 N protein (red) and DAPI (blue). The experiment was conducted twice independently with similar results. e, The ratio of the number of reads related to 2019-nCoV among the total number of virus-related reads in metagenomics analysis of supernatants from Vero E6 cell cultures. f, Virus growth in Vero E6 cells. g, Viral particles in the ultrathin sections were imaged using electron microscopy at 200 kV. The sample was from virus-infected Vero E6 cells. The inset shows the viral particles in an intra-cytosolic vacuole.
Extended Data Fig. 7. Analysis of 2019-nCoV receptor usage.
Determination of virus infectivity in HeLa cells with or without the expression of human APN and DPP4. The expression of ACE2, APN and DPP4 plasmids with S tag were detected using mouse anti-S tag monoclonal antibody. ACE2, APN and DPP4 proteins (green), viral protein (red) and nuclei (blue) are shown. Scale bars, 10 μm.
Comment in
- Potential of large "first generation" human-to-human transmission of 2019-nCoV.
Li X, Zai J, Wang X, Li Y. Li X, et al. J Med Virol. 2020 Apr;92(4):448-454. doi: 10.1002/jmv.25693. Epub 2020 Feb 14. J Med Virol. 2020. PMID: 31997390 Free PMC article. - Calling all coronavirus researchers: keep sharing, stay open.
[No authors listed] [No authors listed] Nature. 2020 Feb;578(7793):7. doi: 10.1038/d41586-020-00307-x. Nature. 2020. PMID: 32020126 No abstract available. - Evolving status of the 2019 novel coronavirus infection: Proposal of conventional serologic assays for disease diagnosis and infection monitoring.
Xiao SY, Wu Y, Liu H. Xiao SY, et al. J Med Virol. 2020 May;92(5):464-467. doi: 10.1002/jmv.25702. Epub 2020 Feb 17. J Med Virol. 2020. PMID: 32031264 Free PMC article. No abstract available. - Novel coronavirus takes flight from bats?
York A. York A. Nat Rev Microbiol. 2020 Apr;18(4):191. doi: 10.1038/s41579-020-0336-9. Nat Rev Microbiol. 2020. PMID: 32051570 Free PMC article. - Chloroquine for the 2019 novel coronavirus SARS-CoV-2.
Colson P, Rolain JM, Raoult D. Colson P, et al. Int J Antimicrob Agents. 2020 Mar;55(3):105923. doi: 10.1016/j.ijantimicag.2020.105923. Epub 2020 Feb 15. Int J Antimicrob Agents. 2020. PMID: 32070753 Free PMC article. No abstract available. - SARS Coronavirus Redux.
Qing E, Gallagher T. Qing E, et al. Trends Immunol. 2020 Apr;41(4):271-273. doi: 10.1016/j.it.2020.02.007. Epub 2020 Mar 12. Trends Immunol. 2020. PMID: 32173256 Free PMC article. - A Unique Protease Cleavage Site Predicted in the Spike Protein of the Novel Pneumonia Coronavirus (2019-nCoV) Potentially Related to Viral Transmissibility.
Wang Q, Qiu Y, Li JY, Zhou ZJ, Liao CH, Ge XY. Wang Q, et al. Virol Sin. 2020 Jun;35(3):337-339. doi: 10.1007/s12250-020-00212-7. Epub 2020 Mar 20. Virol Sin. 2020. PMID: 32198713 Free PMC article. No abstract available. - Inefficiency of Sera from Mice Treated with Pseudotyped SARS-CoV to Neutralize 2019-nCoV Infection.
Liu Z, Xia S, Wang X, Lan Q, Xu W, Wang Q, Jiang S, Lu L. Liu Z, et al. Virol Sin. 2020 Jun;35(3):340-343. doi: 10.1007/s12250-020-00214-5. Epub 2020 Mar 31. Virol Sin. 2020. PMID: 32236815 Free PMC article. No abstract available. - COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome.
Hirano T, Murakami M. Hirano T, et al. Immunity. 2020 May 19;52(5):731-733. doi: 10.1016/j.immuni.2020.04.003. Epub 2020 Apr 22. Immunity. 2020. PMID: 32325025 Free PMC article. - Virus, veritas, vita.
Cheung BMY, Li HL. Cheung BMY, et al. Postgrad Med J. 2020 Jul;96(1137):371-372. doi: 10.1136/postgradmedj-2020-137802. Epub 2020 Apr 28. Postgrad Med J. 2020. PMID: 32345756 No abstract available. - Calling for a COVID-19 One Health Research Coalition.
Amuasi JH, Walzer C, Heymann D, Carabin H, Huong LT, Haines A, Winkler AS. Amuasi JH, et al. Lancet. 2020 May 16;395(10236):1543-1544. doi: 10.1016/S0140-6736(20)31028-X. Epub 2020 May 7. Lancet. 2020. PMID: 32386563 Free PMC article. No abstract available. - [Asthma and COVID-19: a risk population?].
Underner M, Peiffer G, Perriot J, Jaafari N. Underner M, et al. Rev Mal Respir. 2020 Sep;37(7):606-607. doi: 10.1016/j.rmr.2020.05.002. Epub 2020 May 15. Rev Mal Respir. 2020. PMID: 32419737 Free PMC article. French. No abstract available. - The role of Instagram in public health education in COVID-19 in Iran.
Dabbagh A. Dabbagh A. J Clin Anesth. 2020 Oct;65:109887. doi: 10.1016/j.jclinane.2020.109887. Epub 2020 May 20. J Clin Anesth. 2020. PMID: 32454342 Free PMC article. No abstract available. - Isolation and Growth Characteristics of SARS-CoV-2 in Vero Cell.
Yao P, Zhang Y, Sun Y, Gu Y, Xu F, Su B, Chen C, Lu H, Wang D, Yang Z, Niu B, Chen J, Xie L, Chen L, Zhang Y, Wang H, Zhao Y, Guo Y, Ruan J, Zhu Z, Fu Z, Tian D, An Q, Jiang J, Zhu H. Yao P, et al. Virol Sin. 2020 Jun;35(3):348-350. doi: 10.1007/s12250-020-00241-2. Epub 2020 Jun 19. Virol Sin. 2020. PMID: 32562199 Free PMC article. No abstract available. - SARS-CoV-2 Does Not Replicate in Aedes Mosquito Cells nor Present in Field-Caught Mosquitoes from Wuhan.
Xia H, Atoni E, Zhao L, Ren N, Huang D, Pei R, Chen Z, Xiong J, Nyaruaba R, Xiao S, Zhang B, Yuan Z. Xia H, et al. Virol Sin. 2020 Jun;35(3):355-358. doi: 10.1007/s12250-020-00251-0. Epub 2020 Jun 29. Virol Sin. 2020. PMID: 32602045 Free PMC article. No abstract available. - The use of SARS-CoV-2-related coronaviruses from bats and pangolins to polarize mutations in SARS-Cov-2.
Li T, Tang X, Wu C, Yao X, Wang Y, Lu X, Lu J. Li T, et al. Sci China Life Sci. 2020 Oct;63(10):1608-1611. doi: 10.1007/s11427-020-1764-2. Epub 2020 Jul 1. Sci China Life Sci. 2020. PMID: 32621057 Free PMC article. No abstract available. - Indian perspective of remdesivir: A promising COVID-19 drug.
Singh D, Wasan H, Mathur A, Gupta YK. Singh D, et al. Indian J Pharmacol. 2020 May-Jun;52(3):227-228. doi: 10.4103/ijp.IJP_486_20. Epub 2020 Aug 4. Indian J Pharmacol. 2020. PMID: 32874008 Free PMC article. No abstract available. - Genomic Characterization of a New Coronavirus from Migratory Birds in Jiangxi Province of China.
Zhu W, Song W, Fan G, Yang J, Lu S, Jin D, Luo XL, Pu J, Chen H, Xu J. Zhu W, et al. Virol Sin. 2021 Dec;36(6):1656-1659. doi: 10.1007/s12250-021-00402-x. Epub 2021 Jul 8. Virol Sin. 2021. PMID: 34236588 Free PMC article. No abstract available.
Similar articles
- Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.
Wan Y, Shang J, Graham R, Baric RS, Li F. Wan Y, et al. J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article. - Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Lu R, et al. Lancet. 2020 Feb 22;395(10224):565-574. doi: 10.1016/S0140-6736(20)30251-8. Epub 2020 Jan 30. Lancet. 2020. PMID: 32007145 Free PMC article. - Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor.
Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. Ge XY, et al. Nature. 2013 Nov 28;503(7477):535-8. doi: 10.1038/nature12711. Epub 2013 Oct 30. Nature. 2013. PMID: 24172901 Free PMC article. - The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status.
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. Guo YR, et al. Mil Med Res. 2020 Mar 13;7(1):11. doi: 10.1186/s40779-020-00240-0. Mil Med Res. 2020. PMID: 32169119 Free PMC article. Review. - Angiotensin-converting enzyme 2: The old door for new severe acute respiratory syndrome coronavirus 2 infection.
Tan HW, Xu YM, Lau ATY. Tan HW, et al. Rev Med Virol. 2020 Sep;30(5):e2122. doi: 10.1002/rmv.2122. Epub 2020 Jun 30. Rev Med Virol. 2020. PMID: 32602627 Free PMC article. Review.
Cited by
- Detection and Prevalence of Coronaviruses in European Bats: A Systematic Review.
Hemnani M, da Silva PG, Thompson G, Poeta P, Rebelo H, Mesquita JR. Hemnani M, et al. Ecohealth. 2024 Dec;21(2-4):125-140. doi: 10.1007/s10393-024-01688-5. Epub 2024 Nov 23. Ecohealth. 2024. PMID: 39580592 Free PMC article. Review. - COVID-19 vaccines: current and future challenges.
Mohammadi D, Ghasemi M, Manouchehrian N, Zafarmand M, Akbari M, Boroumand AB. Mohammadi D, et al. Front Pharmacol. 2024 Nov 6;15:1434181. doi: 10.3389/fphar.2024.1434181. eCollection 2024. Front Pharmacol. 2024. PMID: 39568586 Free PMC article. Review. - Veterinary medicine under COVID-19: a mixed-methods analysis of student and practitioner experiences in Austria.
Humer E, Winter S, Probst T, Pieh C, Dale R, Brühl D, Neubauer V. Humer E, et al. Front Vet Sci. 2024 Nov 6;11:1460269. doi: 10.3389/fvets.2024.1460269. eCollection 2024. Front Vet Sci. 2024. PMID: 39568480 Free PMC article. - Complete genome sequencing of SARS-CoV-2 strains that were circulating in Uzbekistan over the course of four pandemic waves.
Esonova G, Abdurakhimov A, Ibragimova S, Kurmaeva D, Gulomov J, Mirazimov D, Sohibnazarova K, Abdullaev A, Turdikulova S, Dalimova D. Esonova G, et al. PLoS One. 2024 Nov 19;19(11):e0298940. doi: 10.1371/journal.pone.0298940. eCollection 2024. PLoS One. 2024. PMID: 39561193 Free PMC article. - MetaAll: integrative bioinformatics workflow for analysing clinical metagenomic data.
Bosilj M, Suljič A, Zakotnik S, Slunečko J, Kogoj R, Korva M. Bosilj M, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae597. doi: 10.1093/bib/bbae597. Brief Bioinform. 2024. PMID: 39550223 Free PMC article.
References
- Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous