Transcriptional profiling of human macrophages during infection with Bordetella pertussis - PubMed (original) (raw)
Transcriptional profiling of human macrophages during infection with Bordetella pertussis
Denisa Petráčková et al. RNA Biol. 2020 May.
Erratum in
- Correction.
[No authors listed] [No authors listed] RNA Biol. 2020 Oct;17(10):1520. doi: 10.1080/15476286.2020.1776981. Epub 2020 Jul 3. RNA Biol. 2020. PMID: 32619392 Free PMC article. No abstract available.
Abstract
Bordetella pertussis, a strictly human re-emerging pathogen and the causative agent of whooping cough, exploits a broad variety of virulence factors to establish efficient infection. Here, we used RNA sequencing to analyse the changes in gene expression profiles of human THP-1 macrophages resulting from B. pertussis infection. In parallel, we attempted to determine the changes in intracellular _B. pertussis_-specific transcriptomic profiles resulting from interaction with macrophages. Our analysis revealed that global gene expression profiles in THP-1 macrophages are extensively rewired 6 h post-infection. Among the highly expressed genes, we identified those encoding cytokines, chemokines, and transcription regulators involved in the induction of the M1 and M2 macrophage polarization programmes. Notably, several host genes involved in the control of apoptosis and inflammation which are known to be hijacked by intracellular bacterial pathogens were overexpressed upon infection. Furthermore, in silico analyses identified large temporal changes in expression of specific gene subsets involved in signalling and metabolic pathways. Despite limited numbers of the bacterial reads, we observed reduced expression of majority of virulence factors and upregulation of several transcriptional regulators during infection suggesting that intracellular B. pertussis cells switch from virulent to avirulent phase and actively adapt to intracellular environment, respectively.
Keywords: Bordetella pertussis; host-pathogen interaction; infection; intracellular survival; macrophage.
Figures
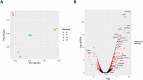
Figure 1.
(A) Principal component analysis was applied to transcriptomic profiles of the uninfected THP-1 macrophages (C; red circles) and _B. pertussis_-infected THP-1 macrophages harvested 2 h (T1, green circles), 6 h (T2, blue circles) and 24 h (T3, cyan circles) pi. Each dot represents an independent biological replicate. (B) Volcano plot showing the global transcriptional changes in the infected THP-1 cells 6 h pi. The red dots represent significantly differentially expressed genes, labelled genes are discussed in this work. The black dots represent nonsignificantly modulated genes.
Figure 2.
Gene Set Enrichment Analysis (GSEA) of GO terms enriched for differentially expressed genes 6 h pi. GSEA was applied to identify enriched GO terms within either downregulated (log2FC < 0) or upregulated (log2FC > 0) gene sets. The bars depict the percentage of DE genes associated with the specific GO term and shades of blue indicate positive log10 of FDR range. Grey bars indicate that this GO term was not significantly enriched within the corresponding gene set. Top 20 enriched gene sets were selected for visualization.
Figure 3.
Enrichment analysis of KEGG pathways enriched for DE genes 6 h pi. Analysis was applied to identify pathways enriched for either downregulated (log2FC < 0) or upregulated (log2FC > 0) DE genes. Top 20 enriched KEGG pathways for each subset of genes were selected for visualization. The legend shows the positive log10 of FDR range (shades of blue) and the number of DE genes in each KEGG pathway expressed by the size of the dots.
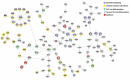
Figure 4.
MSF identification of modulated interacting networks. Networks were generated using data from DE analysis. The network file created by MSF was used to visualize the modulated networks in Cytoscape. The nodes represent the genes and the edges show the direction of interaction. The enrichment within different KEGG pathways (see legend) is shown by the colours around the nodes. The colouring inside the node depicts the range of log fold change for upregulated (shades of red) and downregulated genes (shades of blue).
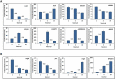
Figure 5.
Validation of the RNA-seq results with quantitative PCR. (A) RT-qPCR analysis was performed to assay the relative expression profiles of IL10, IL23, SOCS3, NFKB1, STAT4, LAMP3, ADORA2A and BIRC3 genes in infected THP-1 macrophages. Relative gene expression was compared between infected and uninfected macrophages harvested 2, 6 and 24 h post-infection (pi). (B) RT-qPCR analysis was performed to assay the relative expression profiles of vag8, prn, vrg6 and BP2871 genes in intracellular B. pertussis cells. Relative gene expression was compared between intracellular and unexposed bacteria 2, 6 and 24 h post-infection (pi). Fold change (FC) values are means (bars) ± standard deviations (error bars) from three biological replicate experiments. Values depicted between the bars denote the fold changes in expression of the specific gene between corresponding time points deduced from RNA-seq results determined 2, 6 and 24 h pi. ND, not determined in the corresponding analysis.
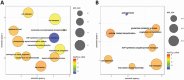
Figure 6.
GO term enrichment analysis of B. pertussis genes modulated during infection. Significantly enriched terms from the domain ‘Biological processes’ were identified within gene sets either upregulated (A) or downregulated (B) between the time points T2 and T1. Results were summarized and visualized by REVIGO as a scatter plot. Circle size encodes number of genes associated with respective category, colours encode the significance level of the enrichment.
Similar articles
- Bordetella pertussis modulates human macrophage defense gene expression.
Valdez HA, Oviedo JM, Gorgojo JP, Lamberti Y, Rodriguez ME. Valdez HA, et al. Pathog Dis. 2016 Aug;74(6):ftw073. doi: 10.1093/femspd/ftw073. Epub 2016 Jul 26. Pathog Dis. 2016. PMID: 27465637 - Transcriptional profiling of Bordetella pertussis reveals requirement of RNA chaperone Hfq for Type III secretion system functionality.
Bibova I, Hot D, Keidel K, Amman F, Slupek S, Cerny O, Gross R, Vecerek B. Bibova I, et al. RNA Biol. 2015;12(2):175-85. doi: 10.1080/15476286.2015.1017237. RNA Biol. 2015. PMID: 25674816 Free PMC article. - Analysis of the In Vivo Transcriptome of Bordetella pertussis during Infection of Mice.
Wong TY, Hall JM, Nowak ES, Boehm DT, Gonyar LA, Hewlett EL, Eby JC, Barbier M, Damron FH. Wong TY, et al. mSphere. 2019 Apr 17;4(2):e00154-19. doi: 10.1128/mSphereDirect.00154-19. mSphere. 2019. PMID: 30996109 Free PMC article. - Environmental sensing mechanisms in Bordetella.
Coote JG. Coote JG. Adv Microb Physiol. 2001;44:141-81. doi: 10.1016/s0065-2911(01)44013-6. Adv Microb Physiol. 2001. PMID: 11407112 Review. - Pertussis: a matter of immune modulation.
de Gouw D, Diavatopoulos DA, Bootsma HJ, Hermans PW, Mooi FR. de Gouw D, et al. FEMS Microbiol Rev. 2011 May;35(3):441-74. doi: 10.1111/j.1574-6976.2010.00257.x. Epub 2011 Jan 5. FEMS Microbiol Rev. 2011. PMID: 21204863 Review.
Cited by
- Eosinophils as drivers of bacterial immunomodulation and persistence.
Parrish KM, Gestal MC. Parrish KM, et al. Infect Immun. 2024 Sep 10;92(9):e0017524. doi: 10.1128/iai.00175-24. Epub 2024 Jul 15. Infect Immun. 2024. PMID: 39007622 Free PMC article. Review. - T3SS chaperone of the CesT family is required for secretion of the anti-sigma factor BtrA in Bordetella pertussis.
Držmíšek J, Petráčková D, Dienstbier A, Čurnová I, Večerek B. Držmíšek J, et al. Emerg Microbes Infect. 2023 Dec;12(2):2272638. doi: 10.1080/22221751.2023.2272638. Epub 2023 Nov 1. Emerg Microbes Infect. 2023. PMID: 37850324 Free PMC article. - Use of an Integrated Multi-Omics Approach To Identify Molecular Mechanisms and Critical Factors Involved in the Pathogenesis of Leptospira.
Kavela S, Vyas P, Cp J, Kushwaha SK, Majumdar SS, Faisal SM. Kavela S, et al. Microbiol Spectr. 2023 Feb 28;11(2):e0313522. doi: 10.1128/spectrum.03135-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36853003 Free PMC article. - Avirulent phenotype promotes Bordetella pertussis adaptation to the intramacrophage environment.
Farman MR, Petráčková D, Kumar D, Držmíšek J, Saha A, Čurnová I, Čapek J, Hejnarová V, Amman F, Hofacker I, Večerek B. Farman MR, et al. Emerg Microbes Infect. 2023 Dec;12(1):e2146536. doi: 10.1080/22221751.2022.2146536. Emerg Microbes Infect. 2023. PMID: 36357372 Free PMC article. - Conquering the host: Bordetella spp. and Pseudomonas aeruginosa molecular regulators in lung infection.
Holban AM, Gregoire CM, Gestal MC. Holban AM, et al. Front Microbiol. 2022 Sep 26;13:983149. doi: 10.3389/fmicb.2022.983149. eCollection 2022. Front Microbiol. 2022. PMID: 36225372 Free PMC article. Review.
References
- WHO . Vaccine preventable deaths and the global immunization vision and strategy, 2006–2015. MMWR Morb Mortal Wkly Rep. 2006;55:511–515. - PubMed
- Cherry JD. The present and future control of pertussis. Clin Infect Dis. 2010;51:663–667. - PubMed
- Raguckas SE, VandenBussche HL, Jacobs C, et al. Pertussis resurgence: diagnosis, treatment, prevention, and beyond. Pharmacotherapy. 2007;27:41–52. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical