Age-related changes to macrophages are detrimental to fracture healing in mice - PubMed (original) (raw)
Age-related changes to macrophages are detrimental to fracture healing in mice
Daniel Clark et al. Aging Cell. 2020 Mar.
Abstract
The elderly population suffers from higher rates of complications during fracture healing that result in increased morbidity and mortality. Inflammatory dysregulation is associated with increased age and is a contributing factor to the myriad of age-related diseases. Therefore, we investigated age-related changes to an important cellular regulator of inflammation, the macrophage, and the impact on fracture healing outcomes. We demonstrated that old mice (24 months) have delayed fracture healing with significantly less bone and more cartilage compared to young mice (3 months). The quantity of infiltrating macrophages into the fracture callus was similar in old and young mice. However, RNA-seq analysis demonstrated distinct differences in the transcriptomes of macrophages derived from the fracture callus of old and young mice, with an up-regulation of M1/pro-inflammatory genes in macrophages from old mice as well as dysregulation of other immune-related genes. Preventing infiltration of the fracture site by macrophages in old mice improved healing outcomes, with significantly more bone in the calluses of treated mice compared to age-matched controls. After preventing infiltration by macrophages, the macrophages remaining within the fracture callus were collected and examined via RNA-seq analysis, and their transcriptome resembled macrophages from young calluses. Taken together, infiltrating macrophages from old mice demonstrate detrimental age-related changes, and depleting infiltrating macrophages can improve fracture healing in old mice.
Keywords: RNA-seq; aging; fracture healing; inflammation; macrophage; osteoimmunology.
© 2020 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
Theodore Miclau has acted as a paid consultant for the following; Amgen, Bone Therapeutics, Arquos, Surrozen, and DePuy, and has received financial or material support from the following: Baxter, Inman Abbott Society, Orthopaedic Research Society, International Combined Orthopaedic Research Societies, Osteosynthesis and Trauma Care Foundation, and AO Foundation/AO Research Institute Advisory Committee. We have obtained an M‐CSFR inhibitor (PLX3397) from Plexxikon Inc. (Berkeley, CA) for this research. All other authors report no conflict of interest.
Figures
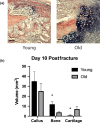
Figure 1
Old mice demonstrate delayed fracture healing compared to young. (a) Representative histological images (HBQ stain) of fracture calluses in old (24 months) and young (3 months) mice (scale bar = 200 µm). Stereological analysis was performed, and the volume of the total callus and the volume of bone and cartilage tissue within the callus were calculated at 10 days after closed tibial fracture (n = 5/group). (b) Old mice demonstrate delayed healing with smaller callus size and significantly less bone and more cartilage (*p < .05)
Figure 2
Immune cell infiltration into the fracture callus is similar in old and young mice. (a) The quantity of B cells, T cells, NKT cells, and NK cells was similar within the fracture callus at days 1, 3, 10, and 14 postfracture measured via flow cytometry in old (n = 5) and young (n = 5) mice. B‐cell quantity at day 10 was the only significant difference between age groups. (b) Macrophages (F4/80+) were the most abundant immune cell analyzed within the fracture callus and demonstrated no significant difference in quantity between young (gray) and old (black) mice at any of the time points analyzed. (c) A subpopulation of macrophages (F4/80+, Ly6C−) was increased in young mice at day 1 compared to old mice (*p < .005)
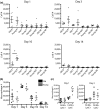
Figure 3
Macrophages from the fracture callus of old mice are transcriptionally distinct from young mice. RNA‐seq analysis of callus macrophages in old (n = 10) and young (n = 11) mice collected at 3 days postfracture. (a) A total of 1,222 genes were significantly differentially expressed in old macrophages compared to young (FDR < 0.1) (red dots). (b) Enriched gene ontology terms associated with the significantly up‐ and down‐regulated transcripts in macrophages from old mice compared to young. (c) Heat map demonstrates unsupervised hierarchical clustering of young (yellow) and old (green) mice based on the differential expression of the 1,222 genes. (d) Hierarchical clustering of young (yellow) and old (green) mice, based on the differential expression of a M1/M2 gene signature, demonstrates a more pro‐inflammatory/M1 gene expression signature in macrophages from old mice compared to young. (e) Principal component analysis of differential gene expression in macrophages from old and young mice demonstrates clustering of young (yellow) mice and a heterogenous spread of old (green) mice across PC1 and PC2
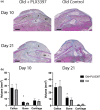
Figure 4
Inhibition of macrophage recruitment improves fracture healing in old mice. (a) Representative histological images (modified Milligan's trichrome stain) of fracture calluses in old mice treated with PLX3397 and age‐matched old controls 10 days and 21 days after closed tibial fracture (n = 6/group) (scale bar = 1mm). B—bone; C—cartilage; BM—bone marrow. Stereological analysis was performed, and the volume of the total callus and the volume of bone and cartilage tissue within the callus were calculated. (b) Old mice treated with PLX3397 demonstrate significantly more bone volume compared to old nontreated mice at both time points (*p < .05)
Figure 5
Improved fracture healing in treated old mice is associated with transcriptionally “younger” macrophages. (a) PLX3397 treatment during fracture healing resulted in significant decrease in macrophage quantity within the fracture callus, as analyzed through flow cytometry. (b) RNA‐seq analysis demonstrates 64 genes were significantly differentially expressed in old macrophages treated with PLX3397 compared to young (red dots) (FDR < 0.1). (c) Hierarchal clustering demonstrates old macrophages treated with PLX3397 (gray) cluster between young (yellow) and old (green) control macrophages. (d) Principal component analysis of differential gene expression in macrophages demonstrates close clustering of young (yellow) mice with old mice treated with PLX3397 (gray)
Similar articles
- T and B cells participate in bone repair by infiltrating the fracture callus in a two-wave fashion.
Könnecke I, Serra A, El Khassawna T, Schlundt C, Schell H, Hauser A, Ellinghaus A, Volk HD, Radbruch A, Duda GN, Schmidt-Bleek K. Könnecke I, et al. Bone. 2014 Jul;64:155-65. doi: 10.1016/j.bone.2014.03.052. Epub 2014 Apr 8. Bone. 2014. PMID: 24721700 - Osteoimmunology of Fracture Healing.
Molitoris KH, Huang M, Baht GS. Molitoris KH, et al. Curr Osteoporos Rep. 2024 Jun;22(3):330-339. doi: 10.1007/s11914-024-00869-z. Epub 2024 Apr 15. Curr Osteoporos Rep. 2024. PMID: 38616228 Free PMC article. Review. - Effects of Aging on Fracture Healing.
Clark D, Nakamura M, Miclau T, Marcucio R. Clark D, et al. Curr Osteoporos Rep. 2017 Dec;15(6):601-608. doi: 10.1007/s11914-017-0413-9. Curr Osteoporos Rep. 2017. PMID: 29143915 Free PMC article. Review. - Effects of aging on the immune and periosteal response to fracture injury.
King JS, Wan M, Kim A, Prabhu S, Novak S, Kalajzic I, Delany AM, Sanjay A. King JS, et al. Bone. 2025 Sep;198:117524. doi: 10.1016/j.bone.2025.117524. Epub 2025 May 15. Bone. 2025. PMID: 40381878 - Complement C3 and C5 deficiency affects fracture healing.
Ehrnthaller C, Huber-Lang M, Nilsson P, Bindl R, Redeker S, Recknagel S, Rapp A, Mollnes T, Amling M, Gebhard F, Ignatius A. Ehrnthaller C, et al. PLoS One. 2013 Nov 18;8(11):e81341. doi: 10.1371/journal.pone.0081341. eCollection 2013. PLoS One. 2013. PMID: 24260573 Free PMC article.
Cited by
- Age-related decrease in periostin expression may be associated with attenuated fracture healing in old mice.
Clark D, Doelling J, Hu D, Miclau T, Nakamura M, Marcucio R. Clark D, et al. J Orthop Res. 2023 May;41(5):1022-1032. doi: 10.1002/jor.25439. Epub 2022 Sep 21. J Orthop Res. 2023. PMID: 36058631 Free PMC article. - Age related effects of selective and non-selective COX-2 inhibitors on bone healing.
Kigera JWM, Gichangi PB, Abdelmalek AKM, Ogeng'o JA. Kigera JWM, et al. J Clin Orthop Trauma. 2022 Jan 12;25:101763. doi: 10.1016/j.jcot.2022.101763. eCollection 2022 Feb. J Clin Orthop Trauma. 2022. PMID: 35211371 Free PMC article. - Immunomodulatory strategies for bone regeneration: A review from the perspective of disease types.
Su N, Villicana C, Yang F. Su N, et al. Biomaterials. 2022 Jul;286:121604. doi: 10.1016/j.biomaterials.2022.121604. Epub 2022 May 25. Biomaterials. 2022. PMID: 35667249 Free PMC article. Review. - Age-related changes to macrophage subpopulations and TREM2 dysregulation characterize attenuated fracture healing in old mice.
Clark D, Brazina S, Miclau T, Park S, Hsieh CL, Nakamura M, Marcucio R. Clark D, et al. Aging Cell. 2024 Sep;23(9):e14212. doi: 10.1111/acel.14212. Epub 2024 Jun 2. Aging Cell. 2024. PMID: 38825965 Free PMC article. - Single-Cell RNA-Sequencing Reveals the Skeletal Cellular Dynamics in Bone Repair and Osteoporosis.
Wu S, Ohba S, Matsushita Y. Wu S, et al. Int J Mol Sci. 2023 Jun 6;24(12):9814. doi: 10.3390/ijms24129814. Int J Mol Sci. 2023. PMID: 37372962 Free PMC article. Review.
References
- Batoon, L. , Millard, S. M. , Wullschleger, M. E. , Preda, C. , Wu, A.‐K. , Kaur, S. , … Pettit, A. R. (2019). CD169 + macrophages are critical for osteoblast maintenance and promote intramembranous and endochondral ossification during bone repair. Biomaterials, 196, 51–66. 10.1016/j.biomaterials.2017.10.033 - DOI - PubMed
- Butowski, N. , Colman, H. , De Groot, J. F. , Omuro, A. M. , Nayak, L. , Wen, P. Y. , … Prados, M. (2016). Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: An Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro‐Oncology, 18, 557–564. 10.1093/neuonc/nov245 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical