The Evolution of Flavonoid Biosynthesis: A Bryophyte Perspective - PubMed (original) (raw)
Review
The Evolution of Flavonoid Biosynthesis: A Bryophyte Perspective
Kevin M Davies et al. Front Plant Sci. 2020.
Abstract
The flavonoid pathway is one of the best characterized specialized metabolite pathways of plants. In angiosperms, the flavonoids have varied roles in assisting with tolerance to abiotic stress and are also key for signaling to pollinators and seed dispersal agents. The pathway is thought to be specific to land plants and to have arisen during the period of land colonization around 550-470 million years ago. In this review we consider current knowledge of the flavonoid pathway in the bryophytes, consisting of the liverworts, hornworts, and mosses. The pathway is less characterized for bryophytes than angiosperms, and the first genetic and molecular studies on bryophytes are finding both commonalities and significant differences in flavonoid biosynthesis and pathway regulation between angiosperms and bryophytes. This includes biosynthetic pathway branches specific to each plant group and the apparent complete absence of flavonoids from the hornworts.
Keywords: anthocyanin; auronidin; dirigent; polyphenol oxidase; transcription factor.
Copyright © 2020 Davies, Jibran, Zhou, Albert, Brummell, Jordan, Bowman and Schwinn.
Figures
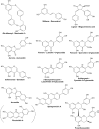
Figure 1
Examples of compounds from phenylpropanoid pathway branches discussed in the text. Bibenzyls, stilbenes, and lignans are phenylpropanoids formed from branches prior to the start of the specific flavonoid pathway branch. The example compounds are from liverworts, angiosperms, and hornworts, respectively. All the other compounds shown are flavonoids.
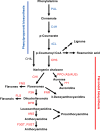
Figure 2
Schematic of a section of the phenylpropanoid pathway leading to the production of major compound groups discussed in the article. Enzyme abbreviations are: PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; CHIL, chalcone isomerase-like; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol 4-reductase; FNR, flavanone reductase; ANS, anthocyanidin synthase; F3GT, flavonoid 3-_O_-glucosyltransferase; A5GT, anthocyanidin 5-_O_-lucosyltransferase; AUS/AS, aureusidin/aurone synthase; FNS, flavone synthase (2OGD = I, Cyp450 = II); FLS, flavonol synthase.
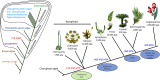
Figure 3
Phylogenetic context of land plants. The phylogenetic tree on the left shows the relationships of the animals, fungi, and plants, including the major divisions within the plants. The tree on the right shows the relationships among the land plants (Embryophyta). The phylogeny of the bryophytes is unresolved, but the current proposal of a sister relationship between liverworts and mosses is shown. MYA, million years ago.
Figure 4
Illustrations of some of the variations in form of the purple-pigmented ventral scales of thalloid liverworts. Left The aquatic form of Ricciocarpos natans (previously Riccia natans) has long ventral scales that extend into the water below the plant. Right Riccia squamata is one of the drought-adapted thalloid liverworts. As the environment dries out it curls over so that the ventral scales encase the thallus. Illustrations are from Lindenberg (1836).
Figure 5
Examples of pigmentation of the thallus and antheridiophores of Marchantia foliacea. Ventral (A, C) and dorsal (B, D) views of a thallus branch. The ventral surface contains the strongly pigmented scales. These extend past the meristematic notch and can be seen from the dorsal surface. The thallus in image (A) shows the strong pigmentation that can also occur in the non-scale cells. Image (D) shows a close up of the meristem region from the dorsal side, with the corresponding ventral view (C) “flipped” vertically to present the same orientation. Image (E) shows the strong pigmentation common for the surface of the antheridiophores.
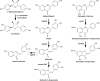
Figure 6
Examples of plant POLYPHENOL OXIDASES with specific activities in specialized metabolism. The biosynthetic activities of three PPOs are shown: (+) Larreatricin hydroxylase, which is part of lignan biosynthesis in the creosote bush (Larrea tridentata), and the AURONE SYNTHASE from Coreopsis grandiflora (_Cg_AUS) and Antirrhinum majus (_Am_AUS).
Figure 7
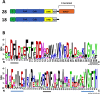
Amino acid sequence features of polyphenol oxidases (PPOs) in Marchantia polymorpha. (A) Number of expressed PPO gene models found and their PPO type. RNA-seq data from Berland et al. (2019) were used to check for expression. The top structure (not to scale) is standard for plant PPOs. Transit/signal peptide (P), copper binding domain TYR : PFAM 00264 (CuA/CuB), PPO1_DWL: PFAM 12142 domain (DWL), tyrosine motif (YxY), PPO1_KFDV : PFAM12143 domain (KFDV). (B) Weblogo display of conserved residues of DWL and KFDV domains based on all gene models having an intact region (generated using
https://weblogo.berkeley.edu/logo.cgi
). DWL, tyrosine, and KFDV core motifs are underlined in black. Residues for the regions identified by Tran et al. (2012) with core motifs of EEEVLV (left) and EFAGSF (right) are underlined in blue.
Similar articles
- Stress, senescence, and specialized metabolites in bryophytes.
Kulshrestha S, Jibran R, van Klink JW, Zhou Y, Brummell DA, Albert NW, Schwinn KE, Chagné D, Landi M, Bowman JL, Davies KM. Kulshrestha S, et al. J Exp Bot. 2022 Jul 16;73(13):4396-4411. doi: 10.1093/jxb/erac085. J Exp Bot. 2022. PMID: 35259256 Free PMC article. Review. - Phylogenomics reveals convergent evolution of red-violet coloration in land plants and the origins of the anthocyanin biosynthetic pathway.
Piatkowski BT, Imwattana K, Tripp EA, Weston DJ, Healey A, Schmutz J, Shaw AJ. Piatkowski BT, et al. Mol Phylogenet Evol. 2020 Oct;151:106904. doi: 10.1016/j.ympev.2020.106904. Epub 2020 Jul 6. Mol Phylogenet Evol. 2020. PMID: 32645485 - Vegetative and reproductive innovations of early land plants: implications for a unified phylogeny.
Renzaglia KS, Duff RJT, Nickrent DL, Garbary DJ. Renzaglia KS, et al. Philos Trans R Soc Lond B Biol Sci. 2000 Jun 29;355(1398):769-93. doi: 10.1098/rstb.2000.0615. Philos Trans R Soc Lond B Biol Sci. 2000. PMID: 10905609 Free PMC article. Review. - Distinct patterns of genome size evolution in each bryophyte lineage are not correlated with whole genome duplication.
Patel N, Budke JM, Bainard J. Patel N, et al. Ann Bot. 2025 Jan 27:mcaf012. doi: 10.1093/aob/mcaf012. Online ahead of print. Ann Bot. 2025. PMID: 39868530 - Genetic analysis of the liverwort Marchantia polymorpha reveals that R2R3MYB activation of flavonoid production in response to abiotic stress is an ancient character in land plants.
Albert NW, Thrimawithana AH, McGhie TK, Clayton WA, Deroles SC, Schwinn KE, Bowman JL, Jordan BR, Davies KM. Albert NW, et al. New Phytol. 2018 Apr;218(2):554-566. doi: 10.1111/nph.15002. Epub 2018 Jan 24. New Phytol. 2018. PMID: 29363139
Cited by
- Untargeted In Silico Compound Classification-A Novel Metabolomics Method to Assess the Chemodiversity in Bryophytes.
Peters K, Balcke G, Kleinenkuhnen N, Treutler H, Neumann S. Peters K, et al. Int J Mol Sci. 2021 Mar 23;22(6):3251. doi: 10.3390/ijms22063251. Int J Mol Sci. 2021. PMID: 33806786 Free PMC article. - Preharvest Application of Phenylalanine Induces Red Color in Mango and Apple Fruit's Skin.
Fanyuk M, Kumar Patel M, Ovadia R, Maurer D, Feygenberg O, Oren-Shamir M, Alkan N. Fanyuk M, et al. Antioxidants (Basel). 2022 Feb 28;11(3):491. doi: 10.3390/antiox11030491. Antioxidants (Basel). 2022. PMID: 35326141 Free PMC article. - Phytochemical Composition and Antibacterial Activity of Barleria albostellata C.B. Clarke Leaf and Stem Extracts.
Gangaram S, Naidoo Y, Dewir YH, Singh M, Lin J, Murthy HN. Gangaram S, et al. Plants (Basel). 2023 Jun 21;12(13):2396. doi: 10.3390/plants12132396. Plants (Basel). 2023. PMID: 37446958 Free PMC article. - Basking in the sun: how mosses photosynthesise and survive in Antarctica.
Yin H, Perera-Castro AV, Randall KL, Turnbull JD, Waterman MJ, Dunn J, Robinson SA. Yin H, et al. Photosynth Res. 2023 Nov;158(2):151-169. doi: 10.1007/s11120-023-01040-y. Epub 2023 Jul 29. Photosynth Res. 2023. PMID: 37515652 Free PMC article. Review. - Molecular characterization of a flavanone 3-hydroxylase gene from citrus fruit reveals its crucial roles in anthocyanin accumulation.
Ma G, Zhang L, Yamamoto R, Kojima N, Yahata M, Kato M. Ma G, et al. BMC Plant Biol. 2023 May 3;23(1):233. doi: 10.1186/s12870-023-04173-3. BMC Plant Biol. 2023. PMID: 37131162 Free PMC article.
References
- Aigner S., Remias D., Karsten U., Holzinger A. (2013). Unusual phenolic compounds contribute to ecophysiological performance in the purple-colored green alga Zygogonium ericetorum (zygnematophyceae, streptophyta) from a high-alpine habitat. J. Phycol. 49, 648–660. 10.1111/jpy.12075 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources