The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2 - PubMed (original) (raw)
The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2
Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. Nat Microbiol. 2020 Apr.
Abstract
The present outbreak of a coronavirus-associated acute respiratory disease called coronavirus disease 19 (COVID-19) is the third documented spillover of an animal coronavirus to humans in only two decades that has resulted in a major epidemic. The Coronaviridae Study Group (CSG) of the International Committee on Taxonomy of Viruses, which is responsible for developing the classification of viruses and taxon nomenclature of the family Coronaviridae, has assessed the placement of the human pathogen, tentatively named 2019-nCoV, within the Coronaviridae. Based on phylogeny, taxonomy and established practice, the CSG recognizes this virus as forming a sister clade to the prototype human and bat severe acute respiratory syndrome coronaviruses (SARS-CoVs) of the species Severe acute respiratory syndrome-related coronavirus, and designates it as SARS-CoV-2. In order to facilitate communication, the CSG proposes to use the following naming convention for individual isolates: SARS-CoV-2/host/location/isolate/date. While the full spectrum of clinical manifestations associated with SARS-CoV-2 infections in humans remains to be determined, the independent zoonotic transmission of SARS-CoV and SARS-CoV-2 highlights the need for studying viruses at the species level to complement research focused on individual pathogenic viruses of immediate significance. This will improve our understanding of virus–host interactions in an ever-changing environment and enhance our preparedness for future outbreaks.
Conflict of interest statement
The authors declare no competing interests.
Figures
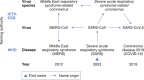
History of coronavirus naming during the three zoonotic outbreaks in relation to virus taxonomy and diseases caused by these viruses.
According to the current international classification of diseases, MERS and SARS are classified as 1D64 and 1D65, respectively.
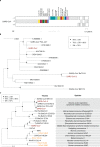
Fig. 1. Taxonomy of selected coronaviruses.
Shown is the full taxonomy of selected coronaviruses in comparison with the taxonomy of humans (the founders of virology and other eminent scientists represent individual human beings for the sake of this comparison), which is given only for categories (ranks) that are shared with the virus taxonomy. Note that these two taxonomies were independently developed using completely different criteria. Although no equivalence is implied, the species of coronaviruses is interpreted sensu stricto as accepted for the species of humans.
Fig. 2. Phylogeny of coronaviruses.
a, Concatenated multiple sequence alignments (MSAs) of the protein domain combination used for phylogenetic and DEmARC analyses of the family Coronaviridae. Shown are the locations of the replicative domains conserved in the ordert Nidovirales in relation to several other ORF1a/b-encoded domains and other major ORFs in the SARS-CoV genome. 5d, 5 domains: nsp5A-3CLpro, two beta-barrel domains of the 3C-like protease; nsp12-NiRAN, nidovirus RdRp-associated nucleotidyltransferase; nsp12-RdRp, RNA-dependent RNA polymerase; nsp13-HEL1 core, superfamily 1 helicase with upstream Zn-binding domain (nsp13-ZBD); nt, nucleotide. b, The maximum-likelihood tree of SARS-CoV was reconstructed by IQ‑TREE v.1.6.1 (ref. ) using 83 sequences with the best fitting evolutionary model. Subsequently, the tree was purged from the most similar sequences and midpoint-rooted. Branch support was estimated using the Shimodaira–Hasegawa (SH)-like approximate likelihood ratio test with 1,000 replicates. GenBank IDs for all viruses except four are shown; SARS-CoV,
AY274119.3
; SARS-CoV-2,
MN908947.3
; SARSr-CoV_BtKY72,
KY352407.1
; SARS-CoV_PC4-227,
AY613950.1
. c, Shown is an IQ‑TREE maximum-likelihood tree of single virus representatives of thirteen species and five representatives of the species Severe acute respiratory syndrome-related coronavirus of the genus Betacoronavirus. The tree is rooted with HCoV-NL63 and HCoV-229E, representing two species of the genus Alphacoronavirus. Purple text highlights zoonotic viruses with varying pathogenicity in humans; orange text highlights common respiratory viruses that circulate in humans. Asterisks indicate two coronavirus species whose demarcations and names are pending approval from the ICTV and, thus, these names are not italicized.
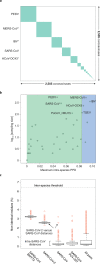
Pairwise distance demarcation of species in the family Coronaviridae.
a, Diagonal matrix of PPDs of 2,505 viruses clustered according to 49 coronavirus species, 39 established and 10 pending or tentative, and ordered from the most to least populous species, from left to right; green and white, PPDs smaller and larger than the inter-species threshold, respectively. Areas of the green squares along the diagonal are proportional to the virus sampling of the respective species, and virus prototypes of the five most sampled species are specified to the left; asterisks indicate species that include viruses whose intra-species PPDs crossed the inter-species threshold (threshold ‘violators’). b, Maximal intra-species PPDs (x axis, linear scale) plotted against virus sampling (y axis, log scale) for 49 species (green dots) of the Coronaviridae. Indicated are the acronyms of virus prototypes of the seven most sampled species. Green and blue plot sections represent intra-species and intra-subgenera PPD ranges. The vertical black line indicates the inter-species threshold. c, Shown are the PDs of non-identical residues (y axis) for four viruses representing three major phylogenetic lineages (clades) of the species Severe acute respiratorysyndrome-related coronavirus (panel b) and all pairs of the 256 viruses of this species (‘all pairs’). The PD values were derived from pairwise distances in the MSA that were calculated using an identity matrix. Panels a and b were adopted from the DEmARC v.1.4 output.
Similar articles
- [Etiology of epidemic outbreaks COVID-19 on Wuhan, Hubei province, Chinese People Republic associated with 2019-nCoV (Nidovirales, Coronaviridae, Coronavirinae, Betacoronavirus, Subgenus Sarbecovirus): lessons of SARS-CoV outbreak.].
Lvov DK, Alkhovsky SV, Kolobukhina LV, Burtseva EI. Lvov DK, et al. Vopr Virusol. 2020;65(1):6-15. doi: 10.36233/0507-4088-2020-65-1-6-15. Vopr Virusol. 2020. PMID: 32496715 Review. Russian. - A pneumonia outbreak associated with a new coronavirus of probable bat origin.
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. Zhou P, et al. Nature. 2020 Mar;579(7798):270-273. doi: 10.1038/s41586-020-2012-7. Epub 2020 Feb 3. Nature. 2020. PMID: 32015507 Free PMC article. - From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes.
Kandeel M, Ibrahim A, Fayez M, Al-Nazawi M. Kandeel M, et al. J Med Virol. 2020 Jun;92(6):660-666. doi: 10.1002/jmv.25754. Epub 2020 Mar 16. J Med Virol. 2020. PMID: 32159237 Free PMC article. - Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses.
Decaro N, Lorusso A. Decaro N, et al. Vet Microbiol. 2020 May;244:108693. doi: 10.1016/j.vetmic.2020.108693. Epub 2020 Apr 14. Vet Microbiol. 2020. PMID: 32402329 Free PMC article. Review. - Susceptibility to SARS, MERS, and COVID-19 from animal health perspective.
Gautam A, Kaphle K, Shrestha B, Phuyal S. Gautam A, et al. Open Vet J. 2020 Aug;10(2):164-177. doi: 10.4314/ovj.v10i2.6. Epub 2020 May 10. Open Vet J. 2020. PMID: 32821661 Free PMC article. Review.
Cited by
- Differential effect on labor force health initiated by the first wave of the COVID-19 in Taiwan.
Yen LC, Su SL, Lee MC, Jiang CJ, Ko PS, Chuang SW, Chen YH, Su W, Lin SY, Cha TL. Yen LC, et al. Medicine (Baltimore). 2024 Nov 8;103(45):e39904. doi: 10.1097/MD.0000000000039904. Medicine (Baltimore). 2024. PMID: 39533618 Free PMC article. - Targeting protein homeostasis with small molecules as a strategy for the development of pan-coronavirus antiviral therapies.
Mao YQ, Jahanshahi S, Malty R, Van Ommen DAJ, Wan Y, Morey TM, Chuang SHW, Pavlova V, Ahmed C, Dahal S, Lin F, Mangos M, Nurtanto J, Song Y, Been T, Christie-Holmes N, Gray-Owen SD, Babu M, Wong AP, Batey RA, Attisano L, Cochrane A, Houry WA. Mao YQ, et al. Commun Biol. 2024 Nov 7;7(1):1460. doi: 10.1038/s42003-024-07143-z. Commun Biol. 2024. PMID: 39511285 Free PMC article. - End-to-end SARS-CoV-2 transmission risks in sport: Current evidence and practical recommendations.
Jones B, Phillips G, Valeriani F, Edwards T, Adams ER, Bonadonna L, Copeland RJ, Cross MJ, Dalton C, Hodgson L, Jimenez A, Kemp SP, Patricios J, Spica VR, Stokes KA, Weed M, Beggs C. Jones B, et al. S Afr J Sports Med. 2021 Jan 15;33(1):v33i1a11210. doi: 10.17159/2078-516X/2021/v33i1a11210. eCollection 2021. S Afr J Sports Med. 2021. PMID: 39498368 Free PMC article. Review. - Identification of a Novel Antiviral Lectin against SARS-CoV-2 Omicron Variant from Shiitake-Mushroom-Derived Vesicle-like Nanoparticles.
Wiggins J, Karim SU, Liu B, Li X, Zhou Y, Bai F, Yu J, Xiang SH. Wiggins J, et al. Viruses. 2024 Sep 30;16(10):1546. doi: 10.3390/v16101546. Viruses. 2024. PMID: 39459880 Free PMC article.
References
- Zheng DP, et al. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. - PubMed
- Adams MJ, et al. 50 years of the International Committee on Taxonomy of Viruses: progress and prospects. Arch. Virol. 2017;162:1441–1446. - PubMed
- Gorbalenya, A. E., Lauber, C. & Siddell, S. Taxonomy of Viruses, in Reference Module in Biomedical Sciences (Elsevier, 2019) 10.1016/B978-0-12-801238-3.99237-7.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous