Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes - PubMed (original) (raw)
. 2020 Apr;17(4):391-394.
doi: 10.1038/s41592-020-0764-5. Epub 2020 Mar 2.
Rafael D Melani 1, Kenneth R Durbin 1, Bon Ikwuagwu 2, Bryan P Early 1, Ryan T Fellers 1, Steven C Beu 3, Vlad Zabrouskov 4, Alexander A Makarov 5, Joshua T Maze 6, Deven L Shinholt 6, Ping F Yip 4, Danielle Tullman-Ercek 2, Michael W Senko 4, Philip D Compton 7, Neil L Kelleher 8
Affiliations
- PMID: 32123391
- PMCID: PMC7131870
- DOI: 10.1038/s41592-020-0764-5
Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes
Jared O Kafader et al. Nat Methods. 2020 Apr.
Abstract
An Orbitrap-based ion analysis procedure determines the direct charge for numerous individual protein ions to generate true mass spectra. This individual ion mass spectrometry (I2MS) method for charge detection enables the characterization of highly complicated mixtures of proteoforms and their complexes in both denatured and native modes of operation, revealing information not obtainable by typical measurements of ensembles of ions.
Conflict of interest statement
COI Statement
V.Z., A.A.M., J.T.M., D.L.S., P.F.Y., and M.W.S. are employees of Thermo Fisher Scientific.
Figures
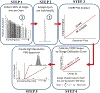
Figure 1.. The five step workflow for the I2MS process.
Includes data acquisition and post-acquisition processing steps that culminate in the plotting of an I2MS spectrum (Step 5). A detailed explanation of each of these five steps is found in the accompanying Online Methods section.
Figure 2.. Deconvolution of Complex Proteoform Mixtures.
Comparison of the number of proteoform assignments possible using either conventional spectral acquisition with an ensemble of ions in a population (a) or the I2MS process (b). This comparative analysis involved directly infusing a mixture of 0 – 30 kDa proteins created by fractionation of whole extracts from HEK-293 human cells. The inset in panel (a) shows an analytical, silver-stained gel, with the fraction analyzed highlighted with a red rectangle. The 3 insets in panel (b) highlight 17 of the 550 total proteoforms assigned, which are listed in Supplemental Table 1 and include their proteoform record numbers (PFRs). The 20–25 kDa region highlighted with a red rectangle in panel (b) corresponds to proteoforms of higher mass proteins in the complex mixture previously unidentified using LC-MS/MS for top-down proteomics.
Figure 3.. Mass Determination of Large Native Complexes.
Crystal structure renderings (a,b) and mass spectra of assembled WT (c,e,g) vs. MINI (d,f,h) MS2 virus-like particles, with approximate masses and capsid diameters of 3.2 vs. 1 MDa and 27 vs.17 nm, respectively. In panels (c) and (d), the standard MS spectra plotted in m/z space results in a loss of charge state resolution due to heterogeneity of cargo inside the capsid of the virus-like particles. However, the corresponding I2MS spectra in panels (e) and (f) allow mass assignment to the distribution of particles even without resolution of the individual charge states (a normal requirement of conventional data produced by electrospray MS). The individual ions assigned to their underlying charge distributions are shown in panels (g) and (h). Mass and resolution (Res.) at full-width of the half-maximum (FWHM) of the peak heights are labeled beside each peak.
Similar articles
- Orbitrap-Based Mass and Charge Analysis of Single Molecules.
Deslignière E, Rolland A, Ebberink EHTM, Yin V, Heck AJR. Deslignière E, et al. Acc Chem Res. 2023 Jun 20;56(12):1458-1468. doi: 10.1021/acs.accounts.3c00079. Epub 2023 Jun 6. Acc Chem Res. 2023. PMID: 37279016 Free PMC article. - Standard Proteoforms and Their Complexes for Native Mass Spectrometry.
Schachner LF, Ives AN, McGee JP, Melani RD, Kafader JO, Compton PD, Patrie SM, Kelleher NL. Schachner LF, et al. J Am Soc Mass Spectrom. 2019 Jul;30(7):1190-1198. doi: 10.1007/s13361-019-02191-w. Epub 2019 Apr 8. J Am Soc Mass Spectrom. 2019. PMID: 30963455 Free PMC article. - Native Proteomics by Capillary Zone Electrophoresis-Mass Spectrometry.
Wang Q, Wang Q, Qi Z, Moeller W, Wysocki VH, Sun L. Wang Q, et al. Angew Chem Int Ed Engl. 2024 Nov 25;63(48):e202408370. doi: 10.1002/anie.202408370. Epub 2024 Oct 24. Angew Chem Int Ed Engl. 2024. PMID: 39196601 - Standardized workflow for multiplexed charge detection mass spectrometry on orbitrap analyzers.
Su P, McGee JP, Hollas MAR, Fellers RT, Durbin KR, Greer JB, Early BP, Yip PF, Zabrouskov V, Srzentić K, Senko MW, Compton PD, Kelleher NL, Kafader JO. Su P, et al. Nat Protoc. 2025 Jun;20(6):1485-1508. doi: 10.1038/s41596-024-01091-y. Epub 2025 Jan 2. Nat Protoc. 2025. PMID: 39747675 Review. - Top-Down Mass Spectrometry: Proteomics to Proteoforms.
Patrie SM. Patrie SM. Adv Exp Med Biol. 2016;919:171-200. doi: 10.1007/978-3-319-41448-5_8. Adv Exp Med Biol. 2016. PMID: 27975217 Review.
Cited by
- Divergent Antibody Repertoires Found for Omicron versus Wuhan SARS-CoV-2 Strains Using Ig-MS.
Forte E, Des Soye BJ, Melani RD, Hollas MAR, Kafader JO, Sha BE, Schneider JR, Kelleher NL. Forte E, et al. J Proteome Res. 2022 Dec 2;21(12):2987-2997. doi: 10.1021/acs.jproteome.2c00514. Epub 2022 Nov 7. J Proteome Res. 2022. PMID: 36343328 Free PMC article. - Not All Arms of IgM Are Equal: Following Hinge-Directed Cleavage by Online Native SEC-Orbitrap-Based CDMS.
Yin V, Deslignière E, Mokiem N, Gazi I, Lood R, de Haas CJC, Rooijakkers SHM, Heck AJR. Yin V, et al. J Am Soc Mass Spectrom. 2024 Jun 5;35(6):1320-1329. doi: 10.1021/jasms.4c00094. Epub 2024 May 20. J Am Soc Mass Spectrom. 2024. PMID: 38767111 Free PMC article. - Proteomes Are of Proteoforms: Embracing the Complexity.
Carbonara K, Andonovski M, Coorssen JR. Carbonara K, et al. Proteomes. 2021 Aug 31;9(3):38. doi: 10.3390/proteomes9030038. Proteomes. 2021. PMID: 34564541 Free PMC article. Review. - Top-Down Stability of Proteins from Rates of Oxidation (TD-SPROX) Approach for Measuring Proteoform-Specific Folding Stability.
Zou Y, Huang CF, Sturrock GR, Kelleher NL, Fitzgerald MC. Zou Y, et al. Anal Chem. 2024 Dec 10;96(49):19597-19604. doi: 10.1021/acs.analchem.4c04469. Epub 2024 Nov 27. Anal Chem. 2024. PMID: 39602376
References
- Aebersold R & Mann M Mass spectrometry-based proteomics. Nature 422, 198–207 (2003). - PubMed
- Elliott AG, Harper CC, Lin H-W & Williams ER Mass, mobility and MSn measurements of single ions using charge detection mass spectrometry. Analyst 142, 2760–2769 (2017). - PubMed
- Keifer DZ, Pierson EE & Jarrold MF Charge detection mass spectrometry: weighing heavier things. Analyst 142, 1654–1671 (2017). - PubMed
- Benner WH A Gated Electrostatic Ion Trap To Repetitiously Measure the Charge and m/z of Large Electrospray Ions. Analytical Chemistry 69, 4162–4168 (1997).
- Schmidt HT, Cederquist H, Jensen J & Fardi A Conetrap: A compact electrostatic ion trap. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 173, 523–527 (2001).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources