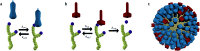Influenza as a molecular walker - PubMed (original) (raw)
Review
. 2019 Nov 14;11(1):27-36.
doi: 10.1039/c9sc05149j. eCollection 2020 Jan 7.
Affiliations
- PMID: 32153750
- PMCID: PMC7021193
- DOI: 10.1039/c9sc05149j
Review
Influenza as a molecular walker
P H Erik Hamming et al. Chem Sci. 2019.
Erratum in
- Correction: Influenza as a molecular walker.
Hamming PHE, Overeem NJ, Huskens J. Hamming PHE, et al. Chem Sci. 2020 Feb 18;11(9):2567. doi: 10.1039/d0sc90015j. Chem Sci. 2020. PMID: 34084421 Free PMC article.
Abstract
The surface of the influenza virus is decorated with the receptor-binding protein hemagglutinin (HA) and the receptor-cleaving enzyme neuraminidase (NA). HA is responsible for host cell recognition, while NA prevents aggregation and entrapment, but the intricate mechanism of how the functions of these glycoproteins cooperate and how they are regulated by mutational responses to environmental pressures remains unclear. Recently, several groups have described the motion of influenza over surfaces and reported that this motion is inhibited by NA inhibitors. We argue that the motion of influenza resembles the motility of artificial receptor-cleaving particles called "molecular spiders". The cleaving of receptors by this type of molecular walkers leads to self-avoiding motion across a surface. When the binding and cleaving rates of molecular spiders are balanced, they move both rapidly and efficiently. The studies of molecular spiders offer new insights into the functional balance of HA and NA, but they do not address the asymmetric distribution of HA and NA on the surface of influenza. We propose that receptor-cleaving molecular walkers could play an important role in the further investigation of the motility of influenza viruses.
This journal is © The Royal Society of Chemistry 2020.
Figures
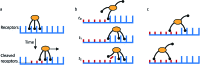
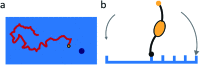
Fig. 1. Mechanism for directional motility in molecular spiders., (a) A molecular spider consists of a rigid body with several legs. The legs bind to receptors on a surface, cleaving them as they go. The legs can bind to a cleaved receptor, but have a lower residence time. (b) The difference in residence time can lead to a bias in movement. At _t_0, the spider is attached at the boundary between fresh and cleaved receptors with one leg. At _t_1, the legs are more likely to be bound to fresh than to cleaved receptors due to the difference in residence times. At _t_2, the first leg detaches and leaves a cleaved receptor behind, shifting the boundary. (c) The spider is either at the boundary and moving with a bias towards fresh receptors, or the spider is in a patch of cleaved receptors where all legs have low residence time and diffusion is fast.
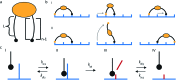
Fig. 2. Main design parameters in molecular spiders. (a) The number of legs (n) of a spider and the length of each leg (L) define the boundary state of the spider. (b) Molecular spiders can walk using an inchworm (I) or hand-over-hand (II) motif. (c) The kinetics of interaction between the substrate and an individual leg.
Fig. 3. (a) Hemagglutinin (HA) is a trimeric protein that binds to sialic acid-terminated glycans in a reversible manner. (b) Neuraminidase (NA) is a tetrameric protein that binds and cleaves sialic acid-terminated glycans. (c) HA and NA are present on the surface of influenza in high copy numbers, approximately in a 6 : 1 ratio.
Fig. 4. Mechanism for virus motion proposed by Sakai et al. (a) Influenza binds tightly to a surface with multiple HA-receptor interactions. (b) NA cleaves receptors, which decreases the number of interactions and initiates motility. (c) A loosely attached virus performs crawling and gliding motions by iterative association and dissociation of HA-receptor interactions, until it reaches a site where it can form multiple interactions and again bind tightly to the surface. Reprinted from ref. 15, with permission from Springer Nature, licensed under CC BY 4.0.
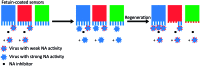
Fig. 5. Motility of influenza is driven by NA activity. De Haan et al. partially blocked receptors on BLI sensors with viruses that had inactive NA and then exposed the sensors to a virus with active NA. One sensor (in blue) was protected with the NA inhibitor oseltamivir carboxylate (OC), one sensor (in red) was only locally blocked by the NA-inactive virus, and one sensor (in green) was left fully unprotected. After regeneration of the sensors, exposure to new virus showed that receptors were cleaved from all unprotected areas. Adapted from ref. 16 with permission from Public Library of Sciences, licensed under CC BY 4.0.
Fig. 6. By labelling HA, NA and cleaved receptors, Vahey and Fletcher showed that the asymmetric organization of HA and NA imparts directional motility in filamentous viruses. Reprinted from ref. 18 with permission from eLife Sciences Publications, licensed under CC BY 4.0.
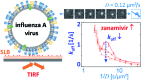
Fig. 7. Block et al. tracked influenza viruses on supported lipid bilayers to quantify the number of interactions with glycolipids. They observed elevated _k_off values for 1/D ∼ 5 s μm–2, which decreased with added NA inhibitor. Reprinted with permission from ref. 17. Copyright 2019 American Chemical Society.
Fig. 8. (a) Self-avoiding walking gives rise to an efficient search pattern to find clathrin-coated pits. (b) The motility of molecular spiders is biased towards a higher density of receptors.
Fig. 9. (a) The structure of the mucus in human airways as proposed by Rubinstein et al. (b) The function of molecular walking of influenza in crossing the mucus. (I) When there is no interaction, particles are repelled by the charged brush. (II) Particles that have an affinity for sialic acid are entrapped. (III) Influenza, which binds and cleaves sialic acid, can walk through the mucus.
P. H. (Erik) Hamming
Nico J. Overeem
Jurriaan Huskens
Similar articles
- Role of Neuraminidase in Influenza A(H7N9) Virus Receptor Binding.
Benton DJ, Wharton SA, Martin SR, McCauley JW. Benton DJ, et al. J Virol. 2017 May 12;91(11):e02293-16. doi: 10.1128/JVI.02293-16. Print 2017 Jun 1. J Virol. 2017. PMID: 28356530 Free PMC article. - Substrate Binding by the Second Sialic Acid-Binding Site of Influenza A Virus N1 Neuraminidase Contributes to Enzymatic Activity.
Du W, Dai M, Li Z, Boons GJ, Peeters B, van Kuppeveld FJM, de Vries E, de Haan CAM. Du W, et al. J Virol. 2018 Sep 26;92(20):e01243-18. doi: 10.1128/JVI.01243-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30089692 Free PMC article. - Biophysical measurement of the balance of influenza a hemagglutinin and neuraminidase activities.
Benton DJ, Martin SR, Wharton SA, McCauley JW. Benton DJ, et al. J Biol Chem. 2015 Mar 6;290(10):6516-21. doi: 10.1074/jbc.M114.622308. Epub 2015 Jan 13. J Biol Chem. 2015. PMID: 25586179 Free PMC article. - Influenza A Virus Hemagglutinin-Neuraminidase-Receptor Balance: Preserving Virus Motility.
de Vries E, Du W, Guo H, de Haan CAM. de Vries E, et al. Trends Microbiol. 2020 Jan;28(1):57-67. doi: 10.1016/j.tim.2019.08.010. Epub 2019 Oct 17. Trends Microbiol. 2020. PMID: 31629602 Free PMC article. Review. - The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase.
Byrd-Leotis L, Cummings RD, Steinhauer DA. Byrd-Leotis L, et al. Int J Mol Sci. 2017 Jul 17;18(7):1541. doi: 10.3390/ijms18071541. Int J Mol Sci. 2017. PMID: 28714909 Free PMC article. Review.
Cited by
- Sialic acids in infection and their potential use in detection and protection against pathogens.
Dedola S, Ahmadipour S, de Andrade P, Baker AN, Boshra AN, Chessa S, Gibson MI, Hernando PJ, Ivanova IM, Lloyd JE, Marín MJ, Munro-Clark AJ, Pergolizzi G, Richards SJ, Ttofi I, Wagstaff BA, Field RA. Dedola S, et al. RSC Chem Biol. 2023 Dec 19;5(3):167-188. doi: 10.1039/d3cb00155e. eCollection 2024 Mar 6. RSC Chem Biol. 2023. PMID: 38456038 Free PMC article. Review. - Motility of an autonomous protein-based artificial motor that operates via a burnt-bridge principle.
Korosec CS, Unksov IN, Surendiran P, Lyttleton R, Curmi PMG, Angstmann CN, Eichhorn R, Linke H, Forde NR. Korosec CS, et al. Nat Commun. 2024 Feb 23;15(1):1511. doi: 10.1038/s41467-024-45570-y. Nat Commun. 2024. PMID: 38396042 Free PMC article. - A novel N95 respirator with chitosan nanoparticles: mechanical, antiviral, microbiological and cytotoxicity evaluations.
Landim MG, Carneiro MLB, Joanitti GA, Anflor CTM, Marinho DD, Rodrigues JFB, de Sousa WJB, Fernandes DO, Souza BF, Ombredane AS, do Nascimento JCF, Felice GJ, Kubota AMA, Barbosa JSC, Ohno JH, Amoah SKS, Pena LJ, Luz GVDS, de Andrade LR, Pinheiro WO, Ribeiro BM, Formiga FR, Fook MVL, Rosa MFF, Peixoto HM, Luiz Carregaro R, Rosa SSRF. Landim MG, et al. Discov Nano. 2023 Sep 21;18(1):118. doi: 10.1186/s11671-023-03892-8. Discov Nano. 2023. PMID: 37733165 Free PMC article. - H3N2 influenza A virus gradually adapts to human-type receptor binding and entry specificity after the start of the 1968 pandemic.
Liu M, Bakker AS, Narimatsu Y, van Kuppeveld FJM, Clausen H, de Haan CAM, de Vries E. Liu M, et al. Proc Natl Acad Sci U S A. 2023 Aug;120(31):e2304992120. doi: 10.1073/pnas.2304992120. Epub 2023 Jul 19. Proc Natl Acad Sci U S A. 2023. PMID: 37467282 Free PMC article. - Receptor Density-Dependent Motility of Influenza Virus Particles on Surface Gradients.
Hamming PHE, Overeem NJ, Diestelhorst K, Fiers T, Tieke M, Vos GM, Boons GPH, van der Vries E, Block S, Huskens J. Hamming PHE, et al. ACS Appl Mater Interfaces. 2023 May 24;15(20):25066-25076. doi: 10.1021/acsami.3c05299. Epub 2023 May 11. ACS Appl Mater Interfaces. 2023. PMID: 37167605 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources