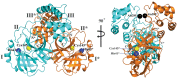Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors - PubMed (original) (raw)
Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors
Linlin Zhang et al. Science. 2020.
Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) is a global health emergency. An attractive drug target among coronaviruses is the main protease (Mpro, also called 3CLpro) because of its essential role in processing the polyproteins that are translated from the viral RNA. We report the x-ray structures of the unliganded SARS-CoV-2 Mpro and its complex with an α-ketoamide inhibitor. This was derived from a previously designed inhibitor but with the P3-P2 amide bond incorporated into a pyridone ring to enhance the half-life of the compound in plasma. On the basis of the unliganded structure, we developed the lead compound into a potent inhibitor of the SARS-CoV-2 Mpro The pharmacokinetic characterization of the optimized inhibitor reveals a pronounced lung tropism and suitability for administration by the inhalative route.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
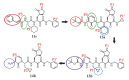
Fig. 1. Chemical structures of α-ketoamide inhibitors 11r, 13a, 13b, and 14b.
Colored ovals and circles highlight the modifications from one development step to the next (see text).
Fig. 2. Three-dimensional structure of SARS-CoV-2 Mpro in two different views.
One protomer of the dimer is shown in light blue, the other one in orange. Domains are labeled by Roman numerals. Amino acid residues of the catalytic site are indicated as yellow spheres for Cys145 and blue spheres for His41. Asterisks mark residues from protomer B (orange). Black spheres indicate the positions of Ala285 for each of the two domains III (see text). Chain termini are labeled N and C for molecule A (light blue) and N* and C* for molecule B (orange).
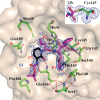
Fig. 3. Compound 13b in the substrate-binding cleft located between domains I and II of the Mpro in the monoclinic crystal form (space group _C_2).
F_obs – F_calc density is shown for the inhibitor (contouring level 3σ). Carbon atoms of the inhibitor are magenta, except in the pyridone ring, which is black; oxygen atoms are red, nitrogens blue, and sulfur yellow. Light blue symbols S_n (n = 1, 2, 3…) indicate the canonical binding pockets for moieties P_n (n = 1, 2, 3…) (red symbols) of the peptidomimetic inhibitor. Hydrogen bonds are indicated by dashed red lines. Note the interaction between Ser1*, the N-terminal residue of molecule B, and Glu166 of molecule A, which is essential for keeping the S1 pocket in the correct shape and the enzyme in the active conformation. Inset: Thiohemiketal formed by the nucleophilic attack of the catalytic cysteine onto the α-carbon of the inhibitor. The stereochemistry of the α-carbon is S. _F_obs − _F_calc density (contoured at 3σ) is shown in blue. See fig. S9 for more details.
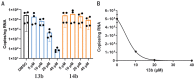
Fig. 4. Compound 13b inhibits SARS-CoV-2 replication in human Calu-3 lung cells.
(A) Calu-3 cells were infected with SARS-CoV-2 using a multiplicity of infection (MOI) of 0.05. Varying amounts (5, 10, 20, or 40 μM) of 13b (blue bars) or 14b (orange bars) were added. DMSO was used as vehicle control (black bar). Total RNA was isolated from cell lysates, and viral RNA content was analyzed by quantitative polymerase chain reaction. Data are means ± SD of two biological experiments with two technical replicates each. (B) For the estimation of the EC50 value of compound 13b against SARS-CoV-2, a dose-response curve was prepared (GraphPad).
Comment in
- Designing of improved drugs for COVID-19: Crystal structure of SARS-CoV-2 main protease Mpro.
Mengist HM, Fan X, Jin T. Mengist HM, et al. Signal Transduct Target Ther. 2020 May 9;5(1):67. doi: 10.1038/s41392-020-0178-y. Signal Transduct Target Ther. 2020. PMID: 32388537 Free PMC article. No abstract available.
Similar articles
- Optimization Rules for SARS-CoV-2 Mpro Antivirals: Ensemble Docking and Exploration of the Coronavirus Protease Active Site.
Stoddard SV, Stoddard SD, Oelkers BK, Fitts K, Whalum K, Whalum K, Hemphill AD, Manikonda J, Martinez LM, Riley EG, Roof CM, Sarwar N, Thomas DM, Ulmer E, Wallace FE, Pandey P, Roy S. Stoddard SV, et al. Viruses. 2020 Aug 26;12(9):942. doi: 10.3390/v12090942. Viruses. 2020. PMID: 32859008 Free PMC article. - Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy.
Goyal B, Goyal D. Goyal B, et al. ACS Comb Sci. 2020 Jun 8;22(6):297-305. doi: 10.1021/acscombsci.0c00058. Epub 2020 May 27. ACS Comb Sci. 2020. PMID: 32402186 Review. - Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Vuong W, Khan MB, Fischer C, Arutyunova E, Lamer T, Shields J, Saffran HA, McKay RT, van Belkum MJ, Joyce MA, Young HS, Tyrrell DL, Vederas JC, Lemieux MJ. Vuong W, et al. Nat Commun. 2020 Aug 27;11(1):4282. doi: 10.1038/s41467-020-18096-2. Nat Commun. 2020. PMID: 32855413 Free PMC article. - Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.
Akaji K, Konno H. Akaji K, et al. Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review. - Discovery of M Protease Inhibitors Encoded by SARS-CoV-2.
Hung HC, Ke YY, Huang SY, Huang PN, Kung YA, Chang TY, Yen KJ, Peng TT, Chang SE, Huang CT, Tsai YR, Wu SH, Lee SJ, Lin JH, Liu BS, Sung WC, Shih SR, Chen CT, Hsu JT. Hung HC, et al. Antimicrob Agents Chemother. 2020 Aug 20;64(9):e00872-20. doi: 10.1128/AAC.00872-20. Print 2020 Aug 20. Antimicrob Agents Chemother. 2020. PMID: 32669265 Free PMC article.
Cited by
- Allostery in homodimeric SARS-CoV-2 main protease.
Fornasier E, Fabbian S, Shehi H, Enderle J, Gatto B, Volpin D, Biondi B, Bellanda M, Giachin G, Sosic A, Battistutta R. Fornasier E, et al. Commun Biol. 2024 Nov 4;7(1):1435. doi: 10.1038/s42003-024-07138-w. Commun Biol. 2024. PMID: 39496839 Free PMC article. - In Silico Identification of Potential Inhibitors of SARS-CoV-2 Main Protease (Mpro).
Hernández-Serda MA, Vázquez-Valadez VH, Aguirre-Vidal P, Markarian NM, Medina-Franco JL, Cardenas-Granados LA, Alarcón-López AY, Martínez-Soriano PA, Velázquez-Sánchez AM, Falfán-Valencia RE, Angeles E, Abrahamyan L. Hernández-Serda MA, et al. Pathogens. 2024 Oct 11;13(10):887. doi: 10.3390/pathogens13100887. Pathogens. 2024. PMID: 39452758 Free PMC article. - Advances in the Search for SARS-CoV-2 Mpro and PLpro Inhibitors.
Diogo MA, Cabral AGT, de Oliveira RB. Diogo MA, et al. Pathogens. 2024 Sep 24;13(10):825. doi: 10.3390/pathogens13100825. Pathogens. 2024. PMID: 39452697 Free PMC article. Review. - Inhibition of the SARS-CoV-2 Non-structural Protein 5 (NSP5) Protease by Nitrosocarbonyl-Bases Small Molecules.
Leusciatti M, Macchi B, Marino-Merlo F, Stefanizzi V, Mastino A, Morra G, Quadrelli P. Leusciatti M, et al. ACS Omega. 2024 Sep 25;9(40):41599-41615. doi: 10.1021/acsomega.4c05480. eCollection 2024 Oct 8. ACS Omega. 2024. PMID: 39398138 Free PMC article. - Mechanism of non-competitive inhibition of the SARS-CoV-2 3CL protease dimerization: Therapeutic and clinical promise of the lichen secondary metabolite perlatolinic acid.
Fagnani L, Bellio P, Di Giulio A, Nazzicone L, Iorio R, Petricca S, Franceschini N, Bertarini L, Tondi D, Celenza G. Fagnani L, et al. Heliyon. 2024 Sep 28;10(19):e38445. doi: 10.1016/j.heliyon.2024.e38445. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39397941 Free PMC article.
References
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
- Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., Haagmans B. L., Lauber C., Leontovich A. M., Neuman B. W., Penzar D., Perlman S., Poon L. L. M., Samborskiy D., Sidorov I. A., Sola I., Ziebuhr J., Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. Nat. Microbiol. 5, 536–544 (2020). 10.1038/s41564-020-0695-z - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Miscellaneous