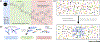Poly(ADP-ribose): A Dynamic Trigger for Biomolecular Condensate Formation - PubMed (original) (raw)
Review
Poly(ADP-ribose): A Dynamic Trigger for Biomolecular Condensate Formation
Anthony K L Leung. Trends Cell Biol. 2020 May.
Abstract
Poly(ADP-ribose) (PAR) is a nucleic acid-like protein modification that can seed the formation of microscopically visible cellular compartments that lack enveloping membranes, recently termed biomolecular condensates. These PAR-mediated condensates are linked to cancer, viral infection, and neurodegeneration. Recent data have shown the therapeutic potential of modulating PAR conjugation (PARylation): PAR polymerase (PARP) inhibitors can modulate the formation and dynamics of these condensates as well as the trafficking of their components - many of which are key disease factors. However, the way in which PARylation facilitates these functions remains unclear, partly because of our lack of understanding of the fundamental parameters of intracellular PARylation, including the sites that are conjugated, PAR chain length and structure, and the physicochemical properties of the conjugates. This review first introduces the role of PARylation in regulating biomolecular condensates, followed by discussion of current knowledge gaps, potential solutions, and therapeutic applications.
Keywords: ADP-ribosylation; biomolecular condensate; liquid–liquid phase separation; poly(ADP-ribose); poly(ADP-ribose) polymerase; poly(ADP-ribose) polymerase inhibitor.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
Fig. 1 |. Poly(ADP-ribose) code (varying sites, length and structure) directs protein interactions within biomolecular condensates.
(A) PAR can be conjugated to different amino acids with varying numbers of ADP-ribose (blue pentagon) and the ADP-ribose can be connected with two possible configurations, resulting in linear and branched chain formation. Analogous to the ubiquitin code, the varying site, length and structure may comprise a PAR code directing biological outcomes. (B) Multivalency can be achieved by multiple PARylated sites from a single protein or by a PAR chain comprising multiple ADP-ribose units for binding to proteins (magenta). Recent data also indicate that the length and structure of PAR are determinants of protein binding (e.g., orange proteins in the illustration require the binding of three ADP-ribose units whereas the red protein binds to the branchpoint of PAR. Therefore, the formation of different PAR may increase the number of multivalent interactions critical for phase separation. (C) PAR can serve a scaffold, where its length and structure specify which client proteins to recruit, resulting in compositional control of biomolecular condensates.
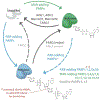
Figure I |
ADP-ribosylation Dynamics
Similar articles
- PARPs and ADP-ribosylation-mediated biomolecular condensates: determinants, dynamics, and disease implications.
Liu H, Pillai M, Leung AKL. Liu H, et al. Trends Biochem Sci. 2025 Mar;50(3):224-241. doi: 10.1016/j.tibs.2024.12.013. Epub 2025 Feb 7. Trends Biochem Sci. 2025. PMID: 39922741 Review. - Regulation of Biomolecular Condensates by Poly(ADP-ribose).
Rhine K, Odeh HM, Shorter J, Myong S. Rhine K, et al. Chem Rev. 2023 Jul 26;123(14):9065-9093. doi: 10.1021/acs.chemrev.2c00851. Epub 2023 Apr 28. Chem Rev. 2023. PMID: 37115110 Free PMC article. Review. - Poly(ADP-ribose): PARadigms and PARadoxes.
Bürkle A, Virág L. Bürkle A, et al. Mol Aspects Med. 2013 Dec;34(6):1046-65. doi: 10.1016/j.mam.2012.12.010. Epub 2013 Jan 2. Mol Aspects Med. 2013. PMID: 23290998 Review. - Poly(ADP-ribose) in Condensates: The PARtnership of Phase Separation and Site-Specific Interactions.
Alemasova EE, Lavrik OI. Alemasova EE, et al. Int J Mol Sci. 2022 Nov 15;23(22):14075. doi: 10.3390/ijms232214075. Int J Mol Sci. 2022. PMID: 36430551 Free PMC article. Review. - Differential and Concordant Roles for Poly(ADP-Ribose) Polymerase 1 and Poly(ADP-Ribose) in Regulating WRN and RECQL5 Activities.
Khadka P, Hsu JK, Veith S, Tadokoro T, Shamanna RA, Mangerich A, Croteau DL, Bohr VA. Khadka P, et al. Mol Cell Biol. 2015 Dec;35(23):3974-89. doi: 10.1128/MCB.00427-15. Epub 2015 Sep 21. Mol Cell Biol. 2015. PMID: 26391948 Free PMC article.
Cited by
- Immediate-Early, Early, and Late Responses to DNA Double Stranded Breaks.
Kieffer SR, Lowndes NF. Kieffer SR, et al. Front Genet. 2022 Jan 31;13:793884. doi: 10.3389/fgene.2022.793884. eCollection 2022. Front Genet. 2022. PMID: 35173769 Free PMC article. Review. - Role of condensates in modulating DNA repair pathways and its implication for chemoresistance.
Dall'Agnese G, Dall'Agnese A, Banani SF, Codrich M, Malfatti MC, Antoniali G, Tell G. Dall'Agnese G, et al. J Biol Chem. 2023 Jun;299(6):104800. doi: 10.1016/j.jbc.2023.104800. Epub 2023 May 9. J Biol Chem. 2023. PMID: 37164156 Free PMC article. Review. - Poly(ADP-Ribosyl) Code Functions.
Maluchenko NV, Koshkina DO, Feofanov AV, Studitsky VM, Kirpichnikov MP. Maluchenko NV, et al. Acta Naturae. 2021 Apr-Jun;13(2):58-69. doi: 10.32607/actanaturae.11089. Acta Naturae. 2021. PMID: 34377556 Free PMC article. - Biochemical Timekeeping Via Reentrant Phase Transitions.
Portz B, Shorter J. Portz B, et al. J Mol Biol. 2021 Jun 11;433(12):166794. doi: 10.1016/j.jmb.2020.166794. Epub 2020 Dec 31. J Mol Biol. 2021. PMID: 33387533 Free PMC article. Review. - PARG is essential for Polθ-mediated DNA end-joining by removing repressive poly-ADP-ribose marks.
Vekariya U, Minakhin L, Chandramouly G, Tyagi M, Kent T, Sullivan-Reed K, Atkins J, Ralph D, Nieborowska-Skorska M, Kukuyan AM, Tang HY, Pomerantz RT, Skorski T. Vekariya U, et al. Nat Commun. 2024 Jul 11;15(1):5822. doi: 10.1038/s41467-024-50158-7. Nat Commun. 2024. PMID: 38987289 Free PMC article.
References
- Nedelsky NB and Taylor JP (2019) Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol 15, 272–286 - PubMed
- Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357, pii: eaaf4382. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases