Protective Microglial Subset in Development, Aging, and Disease: Lessons From Transcriptomic Studies - PubMed (original) (raw)
Review
Protective Microglial Subset in Development, Aging, and Disease: Lessons From Transcriptomic Studies
Anouk Benmamar-Badel et al. Front Immunol. 2020.
Abstract
Microglial heterogeneity has been the topic of much discussion in the scientific community. Elucidation of their plasticity and adaptability to disease states triggered early efforts to characterize microglial subsets. Over time, their phenotypes, and later on their homeostatic signature, were revealed, through the use of increasingly advanced transcriptomic techniques. Recently, an increasing number of these "microglial signatures" have been reported in various homeostatic and disease contexts. Remarkably, many of these states show similar overlapping microglial gene expression patterns, both in homeostasis and in disease or injury. In this review, we integrate information from these studies, and we propose a unique subset, for which we introduce a core signature, based on our own research and reports from the literature. We describe that this subset is found in development and in normal aging as well as in diverse diseases. We discuss the functions of this subset as well as how it is induced.
Keywords: CD11c; CD11c microglia; DAM; heterogeneity; microglia; single cell; subset; transcriptomics.
Copyright © 2020 Benmamar-Badel, Owens and Wlodarczyk.
Figures
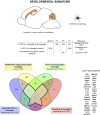
Figure 1
CD11c+ microglia signature in developmental stages. During development, CD11c+ microglia have an amoeboid morphology and localize close to white matter tracts, essentially in the corpus callous and cerebellum. They are present early during embryonic development and their numbers peak between P3 and P7. Comparison of genes upregulated in four studies (24, 35, 36, 70) reveals a common signature for developmental CD11c+ microglia of 39 genes upregulated in at least three of the studies (bold dark outline). Genes shared with the disease signature in Figure 2 are in bold. The Venn diagram was generated using the online tool Venny (72).
Figure 2
CD11c+ microglia signature in disease states. In diseased CNS, CD11c+ microglia adopt an amoeboid, reactive morphology. In AD, they are found surrounding Aβ plaques. Similarly, in MS and ALS models and in injury, they are found around and in the lesions. In glioma, they are found mixed with tumor cells. CD11c+ microglia numbers in diseased CNS vary considerably, ranging from 10 to 50% of all microglia. Comparison of genes upregulated in four studies (, –75) reveals a common signature for CD11c+ disease microglia of 89 genes upregulated in at least three of the studies (bold dark outline). Genes shared with the developmental signature in Figure 1 are in bold. Raw data for the Krasemann study were obtained using the Gene Expression Omnibus Database and the differential gene expression analysis was performed using the DEBrowser package in R (153). The Venn diagram was generated using the online tool Venny (72).
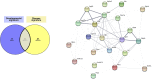
Figure 3
Core CD11c+ microglia signature. Considering similarities between the transcriptomic signatures and functions in the CD11c+ microglia subset in development and in disease, we compared both signatures to obtain a core of genes upregulated in this subset across all conditions. We observe overlap of 20% of the genes between both signatures, corresponding to 22 shared genes. Upon interrogating the STRING database (Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, von Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019 Nov; 47:D607–613.), we observed that the network formed by the proteins corresponding to the genes in the core signature had significantly more interactions than expected from a similar set of random proteins, indicating that these proteins related to the genes in the core signature are at least partially biologically connected. The thickness of the edges linking the different genes is proportional to the strength of the evidence linking the two proteins. The Venn diagram was generated using the online tool Venny (72).
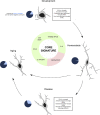
Figure 4
CD11c+ microglia as a subset of microglia present through life and across conditions. Our investigation leads us to believe that CD11c+ microglia represent a subset of microglia characterized by a robust signature of 22 genes expressed by this subset at any age and in various disease states. Emergence of this subset is induced by various factors including signaling through the TREM2–APOE pathway, cell death, IL-4 signaling, and cytokine signaling through CSF1R inducing CCL2, and is inhibited by CD47/SIRPα signaling. In physiological conditions, CD11c+ microglia account for around 15% of all microglia, before contracting to 2% in adulthood and being re-induced by aging at levels similar to development. In disease states, their numbers oscillate between 10 and 50%. We argue that despite the numerous names given to this subset across conditions, it is unique and should be referred to as “CD11c+ microglia”.
Similar articles
- Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer's disease.
Rangaraju S, Dammer EB, Raza SA, Rathakrishnan P, Xiao H, Gao T, Duong DM, Pennington MW, Lah JJ, Seyfried NT, Levey AI. Rangaraju S, et al. Mol Neurodegener. 2018 May 21;13(1):24. doi: 10.1186/s13024-018-0254-8. Mol Neurodegener. 2018. PMID: 29784049 Free PMC article. - The Complementary Role of Morphology in Understanding Microglial Functional Heterogeneity.
Godeanu S, Cătălin B. Godeanu S, et al. Int J Mol Sci. 2025 Apr 17;26(8):3811. doi: 10.3390/ijms26083811. Int J Mol Sci. 2025. PMID: 40332469 Free PMC article. Review. - The Kaleidoscope of Microglial Phenotypes.
Dubbelaar ML, Kracht L, Eggen BJL, Boddeke EWGM. Dubbelaar ML, et al. Front Immunol. 2018 Jul 31;9:1753. doi: 10.3389/fimmu.2018.01753. eCollection 2018. Front Immunol. 2018. PMID: 30108586 Free PMC article. Review. - Microglia heterogeneity along a spatio-temporal axis: More questions than answers.
Silvin A, Ginhoux F. Silvin A, et al. Glia. 2018 Oct;66(10):2045-2057. doi: 10.1002/glia.23458. Epub 2018 Aug 25. Glia. 2018. PMID: 30144321 Review. - Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain.
Xu R, Li X, Boreland AJ, Posyton A, Kwan K, Hart RP, Jiang P. Xu R, et al. Nat Commun. 2020 Mar 27;11(1):1577. doi: 10.1038/s41467-020-15411-9. Nat Commun. 2020. PMID: 32221280 Free PMC article.
Cited by
- Neuronal nuclear calcium signaling suppression of microglial reactivity is mediated by osteoprotegerin after traumatic brain injury.
Fröhlich A, Olde Heuvel F, Rehman R, Krishnamurthy SS, Li S, Li Z, Bayer D, Conquest A, Hagenston AM, Ludolph A, Huber-Lang M, Boeckers T, Knöll B, Morganti-Kossmann MC, Bading H, Roselli F. Fröhlich A, et al. J Neuroinflammation. 2022 Nov 19;19(1):279. doi: 10.1186/s12974-022-02634-4. J Neuroinflammation. 2022. PMID: 36403069 Free PMC article. - miR-214-3p Deficiency Enhances Caspase-1-Dependent Pyroptosis of Microglia in White Matter Injury.
He L, Wei T, Huang Y, Zhang X, Zhu D, Liu H, Wang Z. He L, et al. J Immunol Res. 2022 Aug 22;2022:1642896. doi: 10.1155/2022/1642896. eCollection 2022. J Immunol Res. 2022. PMID: 39262408 Free PMC article. - Absence of miRNA-146a Differentially Alters Microglia Function and Proteome.
Martin NA, Hyrlov KH, Elkjaer ML, Thygesen EK, Wlodarczyk A, Elbaek KJ, Aboo C, Okarmus J, Benedikz E, Reynolds R, Hegedus Z, Stensballe A, Svenningsen ÅF, Owens T, Illes Z. Martin NA, et al. Front Immunol. 2020 Jun 5;11:1110. doi: 10.3389/fimmu.2020.01110. eCollection 2020. Front Immunol. 2020. PMID: 32582192 Free PMC article. - Dendritic Cells and Microglia Have Non-redundant Functions in the Inflamed Brain with Protective Effects of Type 1 cDCs.
Gallizioli M, Miró-Mur F, Otxoa-de-Amezaga A, Cugota R, Salas-Perdomo A, Justicia C, Brait VH, Ruiz-Jaén F, Arbaizar-Rovirosa M, Pedragosa J, Bonfill-Teixidor E, Gelderblom M, Magnus T, Cano E, Del Fresno C, Sancho D, Planas AM. Gallizioli M, et al. Cell Rep. 2020 Oct 20;33(3):108291. doi: 10.1016/j.celrep.2020.108291. Cell Rep. 2020. PMID: 33086061 Free PMC article. - Early microglial response, myelin deterioration and lethality in mice deficient for very long chain ceramide synthesis in oligodendrocytes.
Teo JD, Marian OC, Spiteri AG, Nicholson M, Song H, Khor JXY, McEwen HP, Ge A, Sen MK, Piccio L, Fletcher JL, King NJC, Murray SS, Brüning JC, Don AS. Teo JD, et al. Glia. 2023 Apr;71(4):1120-1141. doi: 10.1002/glia.24329. Epub 2022 Dec 30. Glia. 2023. PMID: 36583573 Free PMC article.
References
- del Río-Hortega P. El “tercer elemento” de los centros nerviosos. I. La microglía en estado normal. II. Intervención de la microglía en los procesos patológicos (células en bastoncito y cuerpos gránulo-adiposos). III. Naturaleza probable de la microglía. Bol Soc Esp Biol. 69–120.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials