SARS-CoV-2 productively infects human gut enterocytes - PubMed (original) (raw)
. 2020 Jul 3;369(6499):50-54.
doi: 10.1126/science.abc1669. Epub 2020 May 1.
Joep Beumer # 2, Jelte van der Vaart # 2, Kèvin Knoops 3, Jens Puschhof 2, Tim I Breugem 1, Raimond B G Ravelli 3, J Paul van Schayck 3, Anna Z Mykytyn 1, Hans Q Duimel 3, Elly van Donselaar 3, Samra Riesebosch 1, Helma J H Kuijpers 3, Debby Schipper 1, Willine J van de Wetering 3, Miranda de Graaf 1, Marion Koopmans 1, Edwin Cuppen 4 5, Peter J Peters 3, Bart L Haagmans # 1, Hans Clevers # 6
Affiliations
- PMID: 32358202
- PMCID: PMC7199907
- DOI: 10.1126/science.abc1669
SARS-CoV-2 productively infects human gut enterocytes
Mart M Lamers et al. Science. 2020.
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can cause coronavirus disease 2019 (COVID-19), an influenza-like disease that is primarily thought to infect the lungs with transmission through the respiratory route. However, clinical evidence suggests that the intestine may present another viral target organ. Indeed, the SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2) is highly expressed on differentiated enterocytes. In human small intestinal organoids (hSIOs), enterocytes were readily infected by SARS-CoV and SARS-CoV-2, as demonstrated by confocal and electron microscopy. Enterocytes produced infectious viral particles, whereas messenger RNA expression analysis of hSIOs revealed induction of a generic viral response program. Therefore, the intestinal epithelium supports SARS-CoV-2 replication, and hSIOs serve as an experimental model for coronavirus infection and biology.
Copyright © 2020, American Association for the Advancement of Science.
Figures
Fig. 1. SARS-CoV and SARS-CoV-2 infect 2D human airway cultures.
(a) Live virus titers can be observed by virus titrations on VeroE6 cells of apical washes at 2, 24, 48, 72 and 96h after infection with SARS-CoV (blue) and SARS-CoV-2 (red). The dotted line indicates the lower limit of detection. Error bars represent SEM. N=4. *P<0.05, **P<0.01, ***P<0.001. (b and c) Immunofluorescent staining of SARS-CoV-2 (b) and SARS-CoV (c) infected differentiated airway cultures. Nucleoprotein (NP) stains viral nucleocapsid (red), which colocalized with the ciliated cell marker AcTUB (green). Goblet cells are identified by MUC5AC (blue). Nuclei are stained with TO-PRO3 (white). Scale bars indicate 20μM. Top panels are side-view while bottom panels are top-view.
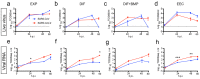
Fig. 2. SARS-CoV and SARS-CoV-2 replicate in hSIOs.
(a to d) Live virus titers can be observed by virus titrations on VeroE6 cells of lysed organoids at 2, 24, 48 and 60h after infection with SARS-CoV (blue) and SARS-CoV-2 (red). Different medium compositions show similar results. (e to h) qPCR analysis targeting the E gene of similar timepoints and medium compositions as (a) to (d). The dotted line indicates the lower limit of detection. Error bars represent SEM. N=3. *P<0.05, **P<0.01, ***P<0.001.
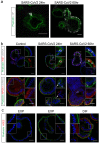
Fig. 3. SARS-CoV-2 infects proliferating cells and enterocytes.
(a) Immunofluorescent staining of SARS-CoV-2-infected intestinal organoids. Nucleoprotein (NP) stains viral capsid. After 24 hours, single virus-infected cells are generally observed in organoids. These small infection clusters spread through the whole organoid after 60 hours. (b) SARS-CoV-2 infects both post-mitotic enterocytes identified by Apolipoprotein A1 (APOA1) and dividing cells that are KI67-positive. Infected cells are visualized by dsRNA staining. Enterocytes are shown in differentiated organoids, and proliferating cells in expanding organoids. Arrows point to APOA1-positive cells. (c) Immunofluorescent staining of ACE2 in intestinal organoids in expansion and differentiation condition. All scale bars are 50 μm.
Fig. 4. Transmission electron microscopy analysis of SARS-CoV-2 infected intestinal organoids.
(a to h) Overview of an intact organoid (a) showing the onset of virus infection (b to d) at different stages of the viral lifecycle, i.e., early double membrane vesicles (DMVs) [(e), asterisk], initial viral production in the Golgi apparatus [(f) and (g)] and complete occupation of virus particles inside the endomembrane system (h). (i to k) Extracellular viruses are observed in the lumen of the organoid (i), and are found at the basal (j) and apical side (k) alongside the microvilli (arrows). Scale bars represent 10 μm (a), 2.5 μm [(b) to (d)], 250 nm [(e), (f), and (h) to (k)] and 100 nm (g). (l to q) Overview of an organoid (l) showing severely infected cells [(m) and (o)], disintegrated cells (o) and stressed cells as evident from the atypical nucleoli (p). Intact cells reveal DMV areas of viral replication [(p), asterisks] and infected Golgi apparatus (q). (r) Extracellular clusters of viruses. Scale bars represent 10 μm (l), 2.5 μm [(m) to (p)] and 250 nm [(p) to (r)]. Data was deposited to the Image Data Resource (
https://idr.openmicroscopy.org
) under accession number idr0083.
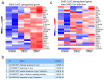
Fig. 5. Transcriptomic analysis of SARS-CoV-2 infected intestinal organoids.
(a) Heatmaps depicting the 25 most significantly enriched genes upon SARS-CoV-2 infection in expanding intestinal organoids. (b) Colored bar represents Z-score of log2 transformed values. GO term enrichment analysis for biological processes of the 50 most significantly up-regulated genes upon SARS-CoV-2 infected in intestinal organoids. (c) Heatmaps depicting the genes from (a) in SARS-CoV infected expanding organoids. Colored bar represents Z-score of log2 transformed values.
Comment in
- Organoids demonstrate gut infection by SARS-CoV-2.
Dickson I. Dickson I. Nat Rev Gastroenterol Hepatol. 2020 Jul;17(7):383. doi: 10.1038/s41575-020-0317-5. Nat Rev Gastroenterol Hepatol. 2020. PMID: 32427981 Free PMC article. No abstract available. - Novel Coronavirus Disease-2019 and the Gastrointestinal Tract: Lessons Learned from Human Organoids.
Jurado-Gomez A, Giraldez MD. Jurado-Gomez A, et al. Gastroenterology. 2020 Dec;159(6):2245-2247. doi: 10.1053/j.gastro.2020.09.039. Epub 2020 Oct 1. Gastroenterology. 2020. PMID: 33010249 Free PMC article. No abstract available. - The mini lungs and other organoids helping to beat COVID.
Mallapaty S. Mallapaty S. Nature. 2021 May;593(7860):492-494. doi: 10.1038/d41586-021-01395-z. Nature. 2021. PMID: 34040221 No abstract available.
Similar articles
- Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.
Busnadiego I, Fernbach S, Pohl MO, Karakus U, Huber M, Trkola A, Stertz S, Hale BG. Busnadiego I, et al. mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article. - Human Intestinal Defensin 5 Inhibits SARS-CoV-2 Invasion by Cloaking ACE2.
Wang C, Wang S, Li D, Wei DQ, Zhao J, Wang J. Wang C, et al. Gastroenterology. 2020 Sep;159(3):1145-1147.e4. doi: 10.1053/j.gastro.2020.05.015. Epub 2020 May 11. Gastroenterology. 2020. PMID: 32437749 Free PMC article. No abstract available. - TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes.
Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ, Ding S. Zang R, et al. Sci Immunol. 2020 May 13;5(47):eabc3582. doi: 10.1126/sciimmunol.abc3582. Sci Immunol. 2020. PMID: 32404436 Free PMC article. - Biological, clinical and epidemiological features of COVID-19, SARS and MERS and AutoDock simulation of ACE2.
Zhang XY, Huang HJ, Zhuang DL, Nasser MI, Yang MH, Zhu P, Zhao MY. Zhang XY, et al. Infect Dis Poverty. 2020 Jul 20;9(1):99. doi: 10.1186/s40249-020-00691-6. Infect Dis Poverty. 2020. PMID: 32690096 Free PMC article. Review. - COVID-19, coronavirus, SARS-CoV-2 and the small bowel.
Mönkemüller K, Fry L, Rickes S. Mönkemüller K, et al. Rev Esp Enferm Dig. 2020 May;112(5):383-388. doi: 10.17235/reed.2020.7137/2020. Rev Esp Enferm Dig. 2020. PMID: 32343593 Review.
Cited by
- Stability and Infectivity of SARS-CoV-2 and Viral RNA in Water, Commercial Beverages, and Bodily Fluids.
Fukuta M, Mao ZQ, Morita K, Moi ML. Fukuta M, et al. Front Microbiol. 2021 May 5;12:667956. doi: 10.3389/fmicb.2021.667956. eCollection 2021. Front Microbiol. 2021. PMID: 34025624 Free PMC article. - JAK inhibitors dampen activation of interferon-stimulated transcription of ACE2 isoforms in human airway epithelial cells.
Lee HK, Jung O, Hennighausen L. Lee HK, et al. Commun Biol. 2021 Jun 2;4(1):654. doi: 10.1038/s42003-021-02167-1. Commun Biol. 2021. PMID: 34079039 Free PMC article. - Mapping the scientific output of organoids for animal and human modeling infectious diseases: a bibliometric assessment.
Yan J, Monlong J, Cougoule C, Lacroix-Lamandé S, Wiedemann A. Yan J, et al. Vet Res. 2024 Jun 26;55(1):81. doi: 10.1186/s13567-024-01333-7. Vet Res. 2024. PMID: 38926765 Free PMC article. - Intestinal Inflammation Modulates the Expression of ACE2 and TMPRSS2 and Potentially Overlaps With the Pathogenesis of SARS-CoV-2-related Disease.
Suárez-Fariñas M, Tokuyama M, Wei G, Huang R, Livanos A, Jha D, Levescot A, Irizar H, Kosoy R, Cording S, Wang W, Losic B, Ungaro RC, Di'Narzo A, Martinez-Delgado G, Suprun M, Corley MJ, Stojmirovic A, Houten SM, Peters L, Curran M, Brodmerkel C, Perrigoue J, Friedman JR, Hao K, Schadt EE, Zhu J, Ko HM, Cho J, Dubinsky MC, Sands BE, Ndhlovu L, Cerf-Bensusan N, Kasarskis A, Colombel JF, Harpaz N, Argmann C, Mehandru S. Suárez-Fariñas M, et al. Gastroenterology. 2021 Jan;160(1):287-301.e20. doi: 10.1053/j.gastro.2020.09.029. Epub 2020 Sep 25. Gastroenterology. 2021. PMID: 32980345 Free PMC article. - Making waves: Wastewater surveillance of SARS-CoV-2 for population-based health management.
Thompson JR, Nancharaiah YV, Gu X, Lee WL, Rajal VB, Haines MB, Girones R, Ng LC, Alm EJ, Wuertz S. Thompson JR, et al. Water Res. 2020 Oct 1;184:116181. doi: 10.1016/j.watres.2020.116181. Epub 2020 Jul 13. Water Res. 2020. PMID: 32707307 Free PMC article.
References
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A. M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A. D. M. E., Schmitz H., Doerr H. W., Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003). 10.1056/NEJMoa030747 - DOI - PubMed
- Guan W. J., Ni Z. Y., Hu Y., Liang W. H., Ou C. Q., He J. X., Liu L., Shan H., Lei C. L., Hui D. S. C., Du B., Li L. J., Zeng G., Yuen K.-Y., Chen R. C., Tang C. L., Wang T., Chen P. Y., Xiang J., Li S. Y., Wang J. L., Liang Z. J., Peng Y. X., Wei L., Liu Y., Hu Y. H., Peng P., Wang J. M., Liu J. Y., Chen Z., Li G., Zheng Z. J., Qiu S. Q., Luo J., Ye C. J., Zhu S. Y., Zhong N. S.; China Medical Treatment Expert Group for Covid-19 , Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). 10.1056/NEJMoa2002032 - DOI - PMC - PubMed
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous