Cell entry mechanisms of SARS-CoV-2 - PubMed (original) (raw)
Cell entry mechanisms of SARS-CoV-2
Jian Shang et al. Proc Natl Acad Sci U S A. 2020.
Abstract
A novel severe acute respiratory syndrome (SARS)-like coronavirus (SARS-CoV-2) is causing the global coronavirus disease 2019 (COVID-19) pandemic. Understanding how SARS-CoV-2 enters human cells is a high priority for deciphering its mystery and curbing its spread. A virus surface spike protein mediates SARS-CoV-2 entry into cells. To fulfill its function, SARS-CoV-2 spike binds to its receptor human ACE2 (hACE2) through its receptor-binding domain (RBD) and is proteolytically activated by human proteases. Here we investigated receptor binding and protease activation of SARS-CoV-2 spike using biochemical and pseudovirus entry assays. Our findings have identified key cell entry mechanisms of SARS-CoV-2. First, SARS-CoV-2 RBD has higher hACE2 binding affinity than SARS-CoV RBD, supporting efficient cell entry. Second, paradoxically, the hACE2 binding affinity of the entire SARS-CoV-2 spike is comparable to or lower than that of SARS-CoV spike, suggesting that SARS-CoV-2 RBD, albeit more potent, is less exposed than SARS-CoV RBD. Third, unlike SARS-CoV, cell entry of SARS-CoV-2 is preactivated by proprotein convertase furin, reducing its dependence on target cell proteases for entry. The high hACE2 binding affinity of the RBD, furin preactivation of the spike, and hidden RBD in the spike potentially allow SARS-CoV-2 to maintain efficient cell entry while evading immune surveillance. These features may contribute to the wide spread of the virus. Successful intervention strategies must target both the potency of SARS-CoV-2 and its evasiveness.
Keywords: ACE2 receptor; COVID-19; SARS-CoV; SARS-CoV-2; proprotein convertase furin.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
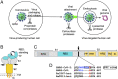
Fig. 1.
PPC motif in SARS-CoV-2 spike protein. (A) Different stages of coronavirus entry where host cellular proteases may activate coronavirus spikes. (B) Schematic drawing of the three-dimensional (3D) structure of coronavirus spike. S1, receptor-binding subunit; S2, membrane fusion subunit; TM, transmembrane anchor; IC, intracellular tail. (C) Schematic drawing of the 1D structure of coronavirus spike. NTD, N-terminal domain. FP (fusion peptide), HR1 (heptad repeat 1), and HR2 (heptad repeat 2) are structural units in coronavirus S2 that function in membrane fusion. (D) Sequence comparison of the spike proteins from SARS-CoV-2, SARS-CoV, and two bat SARS-like coronaviruses in a region at the S1/S2 boundary. Only SARS-CoV-2 spike contains a putative PPC motif—RRAR (residues in the box). The assumed PPC cleavage site is in front of the arginine residue labeled in red. The spike region mutated from SARS-CoV-2 sequence (TNSPRRA) to SARS-CoV sequence (SLL) is labeled in blue. GenBank accession numbers are QHD43416.1 for SARS-CoV-2 spike, AFR58740.1 for SARS-CoV spike, MG916901.1 for bat Rs3367 spike, and QHR63300.2 for bat RaTG13 spike.
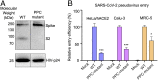
Fig. 2.
Role of PPC motif in SARS-CoV-2 spike-mediated cell entry. (A) Cleavage state of SARS-CoV-2 spike on the surface of pseudoviruses. Packaged SARS-CoV-2 pseudoviruses were subjected to Western blot analysis for detection of the cleavage state of SARS-CoV-2 spike. SARS-CoV-2 spike fragments were detected using anti-C9 antibody targeting the C-terminal C9 tag of the spike protein. (Left) Wild-type (WT) SARS-CoV-2 pseudoviruses. (Right) SARS-CoV-2 pseudoviruses where the PPC motif in the spike protein had been mutated to the corresponding sequence in SARS-CoV spike (see Fig. 1_D_ for details). (B) SARS-CoV-2 pseudovirus entry into three types of target cells. The two types of pseudoviruses correspond to the pseudoviruses in A. Pseudovirus entry efficiency was characterized as luciferase signal accompanying entry. The entry efficiency of wild-type SARS-CoV-2 pseudoviruses was taken as 100%. Error bars indicate SD (n = 4). ***P < 0.001; *P < 0.05.
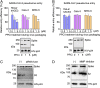
Fig. 3.
Effect of PPCs on SARS-CoV-2 spike-mediated cell entry. (A) SARS-CoV-2 pseudovirus entry into three types of target cells in the presence of PPCi. The pseudoviruses were packaged in the presence of different concentrations of PPCi before they were subjected to cell entry; (-) control: no pseudovirus was added. Also shown is the Western blot result of the corresponding pseudoviruses (packaged in the presence of different concentrations of PPCi). The entry efficiency of SARS-CoV-2 pseudoviruses without any treatment was taken as 100%. Error bars indicate SD (n = 4). ***P < 0.001; **P 0.01; *P < 0.05. (B) SARS-CoV pseudovirus entry into three types of target cells in the presence of PPCi. The experiments were performed in the same way as in A, except that SARS-CoV spike replaced SARS-CoV-2 spike in pseudoviruses. The entry efficiency of SARS-CoV pseudoviruses without any treatment was taken as 100%. (C) Western blot result of SARS-CoV-2 pseudoviruses packaged in cells treated with siRNA. (Left) Pseudoviruses packaged in cells treated with siRNA-negative control. (Right) Pseudoviruses packaged in cells treated with furin-targeting siRNA. (D) Western blot result of SARS-CoV-2 pseudoviruses packaged in cells treated with MMP inhibitor. (Left) Pseudoviruses packaged in cells not treated with MMP inhibitor. (Right) Pseudoviruses packaged in cells treated with MMP inhibitor.
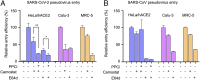
Fig. 4.
Effect of other protease inhibitors on SARS-CoV-2 entry. (A) SARS-CoV-2 pseudovirus entry into three types of target cells in the presence of protease inhibitors. For pseudoviruses treated with PPCi, the pseudoviruses were packaged in the presence of PPCi (5 µM) before they were subjected to cell entry. For pseudoviruses treated with TMPRSS2 inhibitor camostat or lysosomal protease inhibitor E64d, pseudovirus entry was performed in the presence of camostat (50 µM) or E64d (50 µM). The cleavage state of SARS-CoV-2 spike was the same as in Fig. 3_A_ (5 µM PPCi condition). The entry efficiency of SARS-CoV-2 pseudoviruses without any treatment was taken as 100%. Error bars indicate SD (n = 4). ***P < 0.001; *P < 0.05. (B) SARS-CoV pseudovirus entry into three types of target cells. The treatments were done in the same way as in A.
Fig. 5.
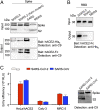
Comparison of receptor binding affinity and cell entry efficiency of SARS-CoV-2 and SARS-CoV. (A) Spike pull-down assay using hACE2 as the bait and cell-associated coronavirus spike molecules as the targets. (Top) Cell-expressed coronavirus spike molecules including SARS-CoV-2 spike, SARS-CoV-2 spike containing a mutant furin site as in Fig. 2_A_, SARS-CoV spike, and MERS-CoV spike. These spike molecules all contain a C-terminal C9 tag. (Middle) Pull-down result using His6-tagged hACE2. (Bottom) Pull-down result using Fc-tagged hACE2. (B) RBD pull-down assay using Fc-tagged hACE2 as the bait and soluble coronavirus RBDs as the targets. These RBD molecules all contain a C-terminal His6 tag. (C) (Left) Entry of SARS-CoV-2 and SARS-CoV pseudoviruses into three types of target cells. (Right) Western blot of SARS-CoV-2 and SARS-CoV pseudoviruses used in the cell entry assay.
Fig. 6.
Summary of cell entry mechanisms of SARS-CoV-2. (A) A schematic view of three unique features of SARS-CoV-2 entry: hidden RBD in the spike for immune evasion, RBD’s high hACE2 binding affinity for efficient entry, and furin preactivation of the spike for enhanced entry into some cells. (B) Implications of the cell entry mechanisms of SARS-CoV-2.
Similar articles
- Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies.
Yi C, Sun X, Ye J, Ding L, Liu M, Yang Z, Lu X, Zhang Y, Ma L, Gu W, Qu A, Xu J, Shi Z, Ling Z, Sun B. Yi C, et al. Cell Mol Immunol. 2020 Jun;17(6):621-630. doi: 10.1038/s41423-020-0458-z. Epub 2020 May 15. Cell Mol Immunol. 2020. PMID: 32415260 Free PMC article. - Structural basis of receptor recognition by SARS-CoV-2.
Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Shang J, et al. Nature. 2020 May;581(7807):221-224. doi: 10.1038/s41586-020-2179-y. Epub 2020 Mar 30. Nature. 2020. PMID: 32225175 Free PMC article. - Enhanced Binding of SARS-CoV-2 Spike Protein to Receptor by Distal Polybasic Cleavage Sites.
Qiao B, Olvera de la Cruz M. Qiao B, et al. ACS Nano. 2020 Aug 25;14(8):10616-10623. doi: 10.1021/acsnano.0c04798. Epub 2020 Aug 4. ACS Nano. 2020. PMID: 32806067 - Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).
Nayak SK. Nayak SK. Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review. - Differences and similarities between SARS-CoV and SARS-CoV-2: spike receptor-binding domain recognition and host cell infection with support of cellular serine proteases.
Rossi GA, Sacco O, Mancino E, Cristiani L, Midulla F. Rossi GA, et al. Infection. 2020 Oct;48(5):665-669. doi: 10.1007/s15010-020-01486-5. Epub 2020 Jul 31. Infection. 2020. PMID: 32737833 Free PMC article. Review.
Cited by
- Brain-wide alterations revealed by spatial transcriptomics and proteomics in COVID-19 infection.
Zhang T, Li Y, Pan L, Sha J, Bailey M, Faure-Kumar E, Williams CK, Wohlschlegel J, Magaki S, Niu C, Lee Y, Su YC, Li X, Vinters HV, Geschwind DH. Zhang T, et al. Nat Aging. 2024 Nov;4(11):1598-1618. doi: 10.1038/s43587-024-00730-z. Epub 2024 Nov 14. Nat Aging. 2024. PMID: 39543407 - Vitamin K2 Protects Against SARS-CoV-2 Envelope Protein-Induced Cytotoxicity in Chronic Myeloid Leukemia Cells and Enhances Imatinib Activity.
Okabe S, Arai Y, Gotoh A. Okabe S, et al. Int J Mol Sci. 2024 Nov 2;25(21):11800. doi: 10.3390/ijms252111800. Int J Mol Sci. 2024. PMID: 39519351 Free PMC article. - Antiseptics: An expeditious third force in the prevention and management of coronavirus diseases.
Okeke KI, Ahamefule CS, Nnabuife OO, Orabueze IN, Iroegbu CU, Egbe KA, Ike AC. Okeke KI, et al. Curr Res Microb Sci. 2024 Oct 16;7:100293. doi: 10.1016/j.crmicr.2024.100293. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39497935 Free PMC article. Review. - COVID-19: a multi-organ perspective.
Guarienti FA, Gonçalves JIB, Gonçalves JB, Antônio Costa Xavier F, Marinowic D, Machado DC. Guarienti FA, et al. Front Cell Infect Microbiol. 2024 Oct 18;14:1425547. doi: 10.3389/fcimb.2024.1425547. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39492990 Free PMC article. Review. - Anti-Inflammatory and Neuroprotective Polyphenols Derived from the European Olive Tree, Olea europaea L., in Long COVID and Other Conditions Involving Cognitive Impairment.
Papadopoulou P, Polissidis A, Kythreoti G, Sagnou M, Stefanatou A, Theoharides TC. Papadopoulou P, et al. Int J Mol Sci. 2024 Oct 14;25(20):11040. doi: 10.3390/ijms252011040. Int J Mol Sci. 2024. PMID: 39456822 Free PMC article. Review.
References
- Lee N. et al. ., A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348, 1986–1994 (2003). - PubMed
- Wu F., et al. , Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv:2020.2003.2030.20047365 (20 April 2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous