Long noncoding RNA lncARSR confers resistance to Adriamycin and promotes osteosarcoma progression - PubMed (original) (raw)
Long noncoding RNA lncARSR confers resistance to Adriamycin and promotes osteosarcoma progression
Peng Shen et al. Cell Death Dis. 2020.
Erratum in
- Correction: Long noncoding RNA lncARSR confers resistance to Adriamycin and promotes osteosarcoma progression.
Shen P, Cheng Y. Shen P, et al. Cell Death Dis. 2021 May 7;12(5):455. doi: 10.1038/s41419-021-03779-5. Cell Death Dis. 2021. PMID: 33963179 Free PMC article. No abstract available.
Abstract
One of the significant challenges for chemotherapy is the appearance of resistance to compounds. Although several signaling pathways have been implicated in the development of Adriamycin (ADM) resistance, mechanisms involved in ADM-resistant osteosarcoma progression remain unknown. The present study attempted to illustrate the role of long noncoding RNA ARSR (lncARSR) in the development of adapted ADM resistance. We found lncARSR overexpressed in the Adriamycin-resistant cell lines U2OS/ADM and MG63/ADM, accompanied with acquired multidrug resistance against to paclitaxel and cisplatin. Overexpression of lncARSR triggered rhodamine 123 efflux and survival, as well as the migration of Adriamycin-resistant cells. Inversely, the depletion of lncARSR promoted rhodamine 123 retention and apoptosis, while reducing the motility of ADM-resistant cells. Further investigation revealed that the upregulation of lncARSR enhanced multidrug resistance-associated protein-1 (MRP1), apoptosis inhibitor Survivin, and matrix metalloproteinase-2 (MMP2) through activating AKT. The reduction of lncARSR overcame the resistance to ADM in U2OS/ADM mouse model. The current study gained novel evidence for understanding the mechanisms underlying adaptive ADM resistance and provided rationales to improve clinical outcomes of refractory osteosarcoma.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
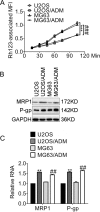
Fig. 1. Resistance profiles of Adriamycin-resistant cells.
a The rhodamine 123-associated median fluorescence intensity (MFI) of the indicated cells. ***P < 0.001, vs. U2OS; ###P < 0.001, vs. MG63. b The expression of the indicated proteins was accessed by immunoblots. c The expression of the indicated genes was accessed by qRT-PCR analysis. **P < 0.01, vs. U2OS; ##P < 0.01, vs. MG63. Data obtained from at least three independent experiments and presented as means plus standard deviation.
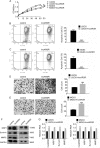
Fig. 2. LncARSR increases in Adriamycin-resistant cells and relates to resistance.
a The heatmap showed the distinct expression of lncRNA in U2OS and U2OS/ADM cells. b The expression of lncARSR in ADM-resistant cells and parental cells was examined by qRT-PCR. ***P < 0.001, vs. U2OS; ###P < 0.001, vs. MG63. c The viability of the indicated cells was detected by MTT assay. Cells were transfected with si-lncARSR or siRNA negative control, and MTT assay was conducted at different time-points. ***P < 0.001, vs. U2OS/ADM or MG63/ADM cells, respectively. d The viability of the indicated cells post-transfection of si-lncARSR or siRNA negative control was accessed by MTT assay. The half-effective inhibition concentrations (IC50) were calculated according to the data obtained from three independent MTT assays. Data obtained from at least three independent experiments and presented as means plus standard deviation.
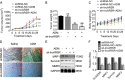
Fig. 3. Overexpression of lncARSR promotes the viability and migration of the parental osteosarcoma cells, while antagonizes apoptosis.
a The viability of cells expressing control or LncRNA ARSR was determined by MTT assay. **P < 0.01, vs. U2OS plus control; ##P < 0.01, vs. MG63 plus control. Apoptosis of (b) U2OS and (c) MG63 cells expressing control or LncARSR was accessed by JC-1 assay following by flow cytometry analysis. ***P < 0.001, vs. control. Migration of (d) U2OS and (e) MG63 cells were detected by transwell assay. ***P < 0.001, vs. control. Scale bar, 200 μm. Magnification, ×100. f The indicated protein expression in U2OS and MG63 cells were analyzed by immunoblots. g The expression of the indicated lncRNA and mRNA in U2OS and MG63 cells were analyzed by qRT-PCR. **P < 0.01, vs. control. Data obtained from at least three independent experiments and presented as means plus standard deviation.
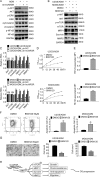
Fig. 4. Reduction of lncARSR impairs the growth, rhodamine 123 efflux, and migration of the Adriamycin-resistant cells while enhancing apoptosis.
a Cell viability of U2OS/ADM or MG63/ADM cells expressing scramble siRNA or si-lncARSR was detected by MTT assay. *P < 0.05, vs. si-NC. b Rh123 retention of U2OS/ADM and MG63/ADM cells expressing si-NC or si-lncARSR was accessed by rhodamine 123 assay and flow cytometry analysis. **P < 0.01, vs. si-NC without ADM. ##P < 0.01, vs. si-NC plus ADM. c The migration of U2OS/ADM cells were detected by transwell assay, respectively. *P < 0.05, vs. si-NC. Scale bar, 200 μm. Magnification, ×100. d The apoptosis of U2OS/ADM or MG63/ADM cells by JC-1 assay followed by flow cytometry analysis. Cells were transfected with siRNA targeting lncARSR. Twenty-four hours later, cells were exposed to 2 µg/ml ADM, and apoptosis was measured at different time-points. *P < 0.05, vs. si-NC; #P < 0.05, vs. si-NC plus ADM. e The indicated genes expression in U2OS/ADM and MG63/ADM cells in the presence or absence of ADM were detected by western blot, separately. f The indicated genes expression in MG63/ADM cells in the presence or absence of ADM were detected by western blot and real-time PCR, separately. si-NC, siRNA negative control. Data obtained from at least three independent experiments and presented as means plus standard deviation.
Fig. 5. The knockdown of lncARSR suppresses tumor growth via promoting sensitivity to ADM.
a Tumor volumes of U2OS/ADM mouse models. Mouse in each group received 6 mg/kg ADM or equal volumes of saline by intraperitoneal injection once per week. ***P < 0.001, vs. shRNA-NC. ns, no significance, vs. shRNA-NC. b Tumor weights of U2OS/ADM mouse models 28 days after treatment with ADM. **P < 0.01, vs. shRNA-NC. ns, no significance, vs. shRNA-NC. ##P < 0.01, vs. sh-lncARSR. c Body weights of U2OS/ADM mouse models. d Representative images for immunohistochemical staining of MRP1. Scale bar, 200 μm. Magnification, ×100. e The indicated protein expression in tumors was accessed by immunoblot. f The expression of the indicated lncRNA and mRNA in tumors was measured by qRT-PCR. *P < 0.05, vs. shRNA-NC. shRNA-NC, shRNA negative control. Data obtained from at least three independent experiments and presented as means plus standard deviation.
Fig. 6. Interference of lncARSR impedes resistance against ADM and retards tumor progression in an Akt-dependent manner.
a The expression of the indicated protein in U2OS/ADM was detected by western blot. b The expression of the indicated protein in ADM-resistant cells was detected by western blot. c The expression of the indicated genes was examined by qRT-PCR. *P < 0.05, vs. parental cells. #P < 0.05, vs. ADM-resistant cells with sh-NC. d The growth of the indicated cells was accessed by MTT assay. *P < 0.05, vs. DMSO. e Rh123 retention of the indicated cells was accessed by rhodamine 123 assay, followed by flow cytometry analysis. *P < 0.05, vs. DMSO. f The apoptosis of the indicated cells was analyzed by JC-1 assay and flow cytometry analysis. *P < 0.05, vs. DMSO. g The migration of the indicated cells was accessed by transwell assay. *P < 0.05, vs. DMSO. Scale bar, 200 μm. Magnification, ×100. h The schematic pathways of lncARSR confers ADM resistance and promotes OS malignancy via activating AKT-mediated cascades. The aberrant expression of lncARSR activates AKT, subsequently enhances mTOR phosphorylation and the expression of MRP1, Survivin, and MMP2, leading to cell growth, acquisition of chemoresistance, survival, and migration. ADM, Adriamycin. OS, osteosarcoma. Cells were exposed to pan-PI3K inhibitor BKM120 50 μM for 24 h before analysis. Data obtained from at least three independent experiments and presented as means plus standard deviation.
Similar articles
- Suppression of adriamycin resistance in osteosarcoma by blocking Wnt/β-catenin signal pathway.
Wu BQ, Cao Y, Bi ZG. Wu BQ, et al. Eur Rev Med Pharmacol Sci. 2017 Jul;21(14):3185-3192. Eur Rev Med Pharmacol Sci. 2017. PMID: 28770967 Retracted. - Long Noncoding RNA lncARSR Promotes Doxorubicin Resistance in Hepatocellular Carcinoma via Modulating PTEN-PI3K/Akt Pathway.
Li Y, Ye Y, Feng B, Qi Y. Li Y, et al. J Cell Biochem. 2017 Dec;118(12):4498-4507. doi: 10.1002/jcb.26107. Epub 2017 Jun 9. J Cell Biochem. 2017. PMID: 28464252 - Cyclin L1 participates in Adriamycin resistance and progression of osteosarcoma via PI3K/AKT-mTOR pathway.
Zhang Y, Zhang T, Chen L, Guo Z, Jiang X. Zhang Y, et al. Aging (Albany NY). 2024 Jun 26;16(14):11208-11223. doi: 10.18632/aging.205972. Epub 2024 Jun 26. Aging (Albany NY). 2024. PMID: 39024509 Free PMC article. - The overexpression of MRP4 is related to multidrug resistance in osteosarcoma cells.
He Z, Hu B, Tang L, Zheng S, Sun Y, Sheng Z, Yao Y, Lin F. He Z, et al. J Cancer Res Ther. 2015 Jan-Mar;11(1):18-23. doi: 10.4103/0973-1482.143334. J Cancer Res Ther. 2015. PMID: 25879330 - Roles of LncRNA ARSR in tumor proliferation, drug resistance, and lipid and cholesterol metabolism.
Li Z, Wang D, Zhu X. Li Z, et al. Clin Transl Oncol. 2025 Apr;27(4):1356-1365. doi: 10.1007/s12094-024-03700-4. Epub 2024 Sep 9. Clin Transl Oncol. 2025. PMID: 39251493 Review.
Cited by
- TPX2 Enhanced the Activation of the HGF/ETS-1 Pathway and Increased the Invasion of Endocrine-Independent Prostate Carcinoma Cells.
Zhou Q, Liu M, Shao T, Xie P, Zhu S, Wang W, Miao Q, Peng J, Zhang P. Zhou Q, et al. Front Oncol. 2021 May 28;11:618540. doi: 10.3389/fonc.2021.618540. eCollection 2021. Front Oncol. 2021. PMID: 34123781 Free PMC article. - LncRNA BCRT1 facilitates osteosarcoma progression via regulating miR-1303/FGF7 axis.
Han G, Guo Q, Ma N, Bi W, Xu M, Jia J, Wang W. Han G, et al. Aging (Albany NY). 2021 Jun 8;13(11):15501-15510. doi: 10.18632/aging.203106. Epub 2021 Jun 8. Aging (Albany NY). 2021. PMID: 34102610 Free PMC article. - Long Noncoding RNAs and Circular RNAs Regulate AKT and Its Effectors to Control Cell Functions of Cancer Cells.
Tang JY, Chuang YT, Shiau JP, Yang KH, Chang FR, Hou MF, Farooqi AA, Chang HW. Tang JY, et al. Cells. 2022 Sep 20;11(19):2940. doi: 10.3390/cells11192940. Cells. 2022. PMID: 36230902 Free PMC article. Review. - A four-lncRNA risk signature for prognostic prediction of osteosarcoma.
Liu H, Chen C, Liu L, Wang Z. Liu H, et al. Front Genet. 2023 Jan 4;13:1081478. doi: 10.3389/fgene.2022.1081478. eCollection 2022. Front Genet. 2023. PMID: 36685868 Free PMC article. - MicroRNA-181a-5p Promotes Osteosarcoma Progression via PTEN/AKT Pathway.
Sun C, Chen C, Chen Z, Guo J, Yu ZH, Qian W, Ai F, Xiao L, Guo X. Sun C, et al. Anal Cell Pathol (Amst). 2022 Mar 8;2022:3421600. doi: 10.1155/2022/3421600. eCollection 2022. Anal Cell Pathol (Amst). 2022. PMID: 35310933 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous