Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients - PubMed (original) (raw)
. 2020 Jun 10;27(6):883-890.e2.
doi: 10.1016/j.chom.2020.04.017. Epub 2020 May 4.
Lili Ren 2, Li Zhang 3, Jiaxin Zhong 4, Yan Xiao 5, Zhilong Jia 6, Li Guo 5, Jing Yang 4, Chun Wang 4, Shuai Jiang 3, Donghong Yang 7, Guoliang Zhang 8, Hongru Li 9, Fuhui Chen 10, Yu Xu 7, Mingwei Chen 11, Zhancheng Gao 7, Jian Yang 5, Jie Dong 5, Bo Liu 5, Xiannian Zhang 12, Weidong Wang 6, Kunlun He 6, Qi Jin 5, Mingkun Li 13, Jianwei Wang 14
Affiliations
- PMID: 32407669
- PMCID: PMC7196896
- DOI: 10.1016/j.chom.2020.04.017
Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients
Zhuo Zhou et al. Cell Host Microbe. 2020.
Abstract
The outbreaks of 2019 novel coronavirus disease (COVID-19) caused by SARS-CoV-2 infection have posed a severe threat to global public health. It is unclear how the human immune system responds to this infection. Here, we used metatranscriptomic sequencing to profile immune signatures in the bronchoalveolar lavage fluid of eight COVID-19 cases. The expression of proinflammatory genes, especially chemokines, was markedly elevated in COVID-19 cases compared to community-acquired pneumonia patients and healthy controls, suggesting that SARS-CoV-2 infection causes hypercytokinemia. Compared to SARS-CoV, which is thought to induce inadequate interferon (IFN) responses, SARS-CoV-2 robustly triggered expression of numerous IFN-stimulated genes (ISGs). These ISGs exhibit immunopathogenic potential, with overrepresentation of genes involved in inflammation. The transcriptome data was also used to estimate immune cell populations, revealing increases in activated dendritic cells and neutrophils. Collectively, these host responses to SARS-CoV-2 infection could further our understanding of disease pathogenesis and point toward antiviral strategies.
Keywords: Bronchoalveolar lavage fluid; COVID-19; Chemokines; Coronavirus; Hypercytokinemia; Innate immune response; Interferon response; Interferon-stimulated genes; Metatranscriptomic sequencing; SARS-CoV-2.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
Graphical abstract
Figure 1
Analysis of DEGs in BALF of COVID-19 and CAP Patients Compared to Healthy Controls (A) Volcano plot of DEGs comparing SARS2 versus Healthy (SARS2-H), Virus-like CAP versus Healthy (Vir-H), and Non-viral CAP versus Healthy (NonVir-H). The names of DEGs with the top 20 absolute FC are shown. (B) PCA loading plot based on all DEGs. Autoscaling of data was performed. (C) Functional enrichment analysis of DEGs with IPA. Asterisks (∗) indicate q-values < 0.05 and absolute _Z_ score ≥ 1. (D) PPI network of upregulated DEGs in SARS2 comparing to Healthy. Each node represents a protein, and interactions with confidence score > 0.9 are presented. See also Figure S2.
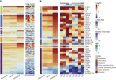
Figure 2
Cytokine-Related Gene Expressions in COVID-19 and CAP Patients (A) Heatmap of 218 genes encoding cytokines and receptors. (B) Heatmap of DEGs encoding cytokines and receptors. SARS2 samples (n = 8) were ordered by days after symptom onset (DSO) in the right panel of (A) and (B). Asterisks (∗) indicate significant DEGs (absolute log2FC ≥ 2, q-value < 0.05). Relative viral reads (calculated by the ratio of SARS-CoV-2 reads to human reads) and the ratios of IL1B to IL1RN are shown in (A) and (B).
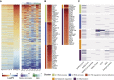
Figure 3
Expression of ISGs in COVID-19 and CAP Patients (A) Heatmap of 628 ISGs. SARS2 samples (n = 8) were ordered by days after symptom onset (DSO) in the right panel. (B) Heatmap of 83 upregulated ISGs in SARS2 comparing to Healthy. Asterisks (∗) indicate significant DEGs (absolute log2FC ≥ 2, q-value < 0.05). (C) Upregulated ISGs in SARS-CoV-2 infection identified in this study, as well as in SARS-CoV and other viral infections (see Table S4). In (B) and (C), ISGs were assigned into five biclusters.
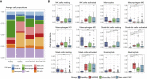
Figure 4
Composition of Immune Cells in BALF Predicted from Transcriptome Data (A) The proportion of nine major immune cell types. (B) The proportion of 12 innate-immunity-related cell subtypes. Asterisks represent significant differences between groups (∗q-value < 0.05, ∗∗q-value < 0.01, ∗∗∗q-value < 0.001, Mann-Whitney test). See also Figure S3.
Similar articles
- Immune and Metabolic Signatures of COVID-19 Revealed by Transcriptomics Data Reuse.
Gardinassi LG, Souza COS, Sales-Campos H, Fonseca SG. Gardinassi LG, et al. Front Immunol. 2020 Jun 26;11:1636. doi: 10.3389/fimmu.2020.01636. eCollection 2020. Front Immunol. 2020. PMID: 32670298 Free PMC article. - Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures.
Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, Aoued H, Tharp GM, Pellegrini KL, Manfredi C, Sorscher E, Mainou B, Lobby JL, Kohlmeier JE, Lowen AC, Shi PY, Menachery VD, Anderson LJ, Grakoui A, Bosinger SE, Suthar MS. Vanderheiden A, et al. J Virol. 2020 Sep 15;94(19):e00985-20. doi: 10.1128/JVI.00985-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32699094 Free PMC article. - Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19.
Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. Liao M, et al. Nat Med. 2020 Jun;26(6):842-844. doi: 10.1038/s41591-020-0901-9. Epub 2020 May 12. Nat Med. 2020. PMID: 32398875 - Covid-19: Perspectives on Innate Immune Evasion.
Taefehshokr N, Taefehshokr S, Hemmat N, Heit B. Taefehshokr N, et al. Front Immunol. 2020 Sep 30;11:580641. doi: 10.3389/fimmu.2020.580641. eCollection 2020. Front Immunol. 2020. PMID: 33101306 Free PMC article. Review. - Prevention and treatment of COVID-19 disease by controlled modulation of innate immunity.
Schijns V, Lavelle EC. Schijns V, et al. Eur J Immunol. 2020 Jul;50(7):932-938. doi: 10.1002/eji.202048693. Epub 2020 Jun 15. Eur J Immunol. 2020. PMID: 32438473 Free PMC article. Review.
Cited by
- Virome analysis provides new insights into the pathogenesis mechanism and treatment of SLE disease.
Wu Y, Zhang Z, Wang X, Liu X, Qiu Y, Ge X, Miao Z, Meng X, Peng Y. Wu Y, et al. Front Cell Infect Microbiol. 2024 Oct 24;14:1484529. doi: 10.3389/fcimb.2024.1484529. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39512588 Free PMC article. - The Potential of Glucosinolates and Their Hydrolysis Products as Inhibitors of Cytokine Storms.
Ochar K, Iwar K, Nair VD, Chung YJ, Ha BK, Kim SH. Ochar K, et al. Molecules. 2024 Oct 11;29(20):4826. doi: 10.3390/molecules29204826. Molecules. 2024. PMID: 39459194 Free PMC article. Review. - Expression of interferon-stimulated genes, but not polymorphisms in the interferon α/β receptor 2 gene, is associated with coronavirus disease 2019 mortality.
Hamidah B, Pakpahan C, Wulandari L, Tinduh D, Wibawa T, Prakoeswa CRS, Oceandy D. Hamidah B, et al. Heliyon. 2024 Oct 5;10(19):e39002. doi: 10.1016/j.heliyon.2024.e39002. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39435115 Free PMC article. - Omicron XBB.1.5 subvariant causes severe pulmonary disease in K18-hACE-2 mice.
Elsharkawy A, Stone S, Guglani A, Patterson LD, Ge C, Dim C, Miano JM, Kumar M. Elsharkawy A, et al. Front Microbiol. 2024 Oct 2;15:1466980. doi: 10.3389/fmicb.2024.1466980. eCollection 2024. Front Microbiol. 2024. PMID: 39417078 Free PMC article. - Future applications of host direct therapies for infectious disease treatment.
Thom RE, D'Elia RV. Thom RE, et al. Front Immunol. 2024 Oct 1;15:1436557. doi: 10.3389/fimmu.2024.1436557. eCollection 2024. Front Immunol. 2024. PMID: 39411713 Free PMC article. Review.
References
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B. 1995;57:289–300.
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Canadian SARS Research Network Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. - PMC - PubMed
- Chaussabel D. Assessment of immune status using blood transcriptomics and potential implications for global health. Semin. Immunol. 2015;27:58–66. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous