Transcriptome Profiling Reveals Indoxyl Sulfate Should Be Culpable of Impaired T Cell Function in Chronic Kidney Disease - PubMed (original) (raw)
Transcriptome Profiling Reveals Indoxyl Sulfate Should Be Culpable of Impaired T Cell Function in Chronic Kidney Disease
Fangfang Xiang et al. Front Med (Lausanne). 2020.
Abstract
Introduction: Chronic inflammation and immune system dysfunction have been evaluated as major factors in the pathogenesis of chronic kidney disease (CKD), contributing to the high mortality rates observed in these populations. Uremic toxins seem to be the potential "missing link." Indoxyl sulfate (IS) is one of the protein-bound renal toxins. It participates in multiple pathologies of CKD complications, yet its effect on immune cell has not been studied. This study aimed to explore the genome-wide expression profile in human peripheral blood T cells under stimulation by IS. Methods: In this study, we employed RNA-sequencing transcriptome profiling to identify differentially expressed genes (DEGs) responding to IS stimulation in human peripheral T cells in vitro. Flow cytometry and western blot were used to verify the discovery in RNA-sequencing analysis. Results: Our results yielded a total of 5129 DEGs that were at least twofold up-regulated or down-regulated significantly by IS stimulation and half of them were concentration-specific. Analysis of T cell functional markers revealed a quite different transcription profile under various IS concentration. Transcription factors analysis showed the similar pattern. Aryl hydrocarbon receptor (AhR) target genes CYP1A1, CYP1B1, NQO1, and AhRR were up-regulated by IS stimulation. Pro-inflammatory genes TNF-α and IFN-γ were up-regulated as verified by flow cytometry analysis. DNA damage was induced by IS stimulation as confirmed by elevated protein level of p-ATM, p-ATR, p-BRCA1, and p-p53 in T cells. Conclusion: The toxicity of IS to T cells could be an important source of chronic inflammation in CKD patients. As an endogenous ligand of AhR, IS may influence multiple biological functions of T cells including inflammatory response and cell cycle regulation. Further researches are required to promulgate the underling mechanism and explore effective method of reserving T cell function in CKD.
Keywords: RNA-sequencing; T cell; aryl hydrocarbon receptor; chronic kidney disease; indoxyl sulfate.
Copyright © 2020 Xiang, Cao, Shen, Chen, Guo, Ding and Zou.
Figures
Figure 1
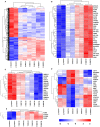
T cell purity and global patterns of RNA-seq profiling. (A) The purity of T cells confirmed by flow cytometry; (B) Hierarchical clustering based on Pearson correlation coefficients among samples in IS groups; (C) Volcano plot showed the numbers of up and down regulated DEGs in each IS groups compared to the control group; (D) Veen figure showed the common and different DEGs in each IS groups.
Figure 2
Hierarchical cluster analysis of patterns of DEGs. (A) 1,332 common DEGs in all IS groups compared with the control groups; (B) 29 DEGs up-regulated by IS simulation in a concentration dependent manner; (C) 15 DEGs down-regulated by IS stimulation in a concentration dependent manner; (D) 20 DEGs up-regulated or down-regulated at 200 μM but reversely regulated when IS concentration was higher; (E) AhR target genes CYP1A1, CYP1B1, NQO1, and AhRR were up-regulated by IS stimulation.
Figure 3
The distribution of TF families. The x-axis represents different TF families (gene names were presented directly if there was only one TF in this TF family), the y-axis represents the percentage of corresponding TF family in total differentially expressed TFs.
Figure 4
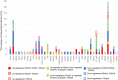
FPKM of TFs involved in T cell differentiation in each IS group. The x-axis represents FPKM, the y-axis represents gene name and IS concentration. *corrected P < 0.05 compared with control group; +corrected P < 0.05 compared with IS 200 μM group; #corrected P < 0.05 compared with IS 1,000 μM group.
Figure 5
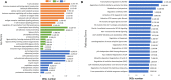
Barplots of significantly enriched terms. (A) GO enrichment and (B) Reactome enrichment terms under corrected P < 0.001. The x-axis represents differentially expressed genes number, the y-axis represents GO or Reactome pathway terms; the numbers in the plot are the corrected _P_-values.
Figure 6
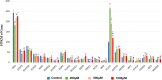
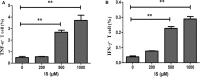
Secretion of TNF–α and IFN-γ in T cells by IS intervention. (A) Secretion of TNF–α was significantly elevated in T cells in IS 500 μM group and IS 1,000 μM group; (B) Secretion of IFN-γ was significantly elevated in T cell in IS 500μM group and IS 1,000 μM group. **P < 0.001 compared with the control group.
Figure 7
Expression of DDR protein in T cells by IS intervention. Protein level of p-ATM, p-ATR, p-p53 and p-BRCA1 were significantly higher in IS 500 μM group compared with the control group. Protein level of p-BRCA1 were also increased in IS 200 μM group. AhR were also up-regulated in IS 200 μM and IS 500 μM group. *P < 0.05 compared with the control group.
Similar articles
- Aryl hydrocarbon receptor is activated in patients and mice with chronic kidney disease.
Dou L, Poitevin S, Sallée M, Addi T, Gondouin B, McKay N, Denison MS, Jourde-Chiche N, Duval-Sabatier A, Cerini C, Brunet P, Dignat-George F, Burtey S. Dou L, et al. Kidney Int. 2018 Apr;93(4):986-999. doi: 10.1016/j.kint.2017.11.010. Epub 2018 Feb 1. Kidney Int. 2018. PMID: 29395338 - Crucial Role of the Aryl Hydrocarbon Receptor (AhR) in Indoxyl Sulfate-Induced Vascular Inflammation.
Ito S, Osaka M, Edamatsu T, Itoh Y, Yoshida M. Ito S, et al. J Atheroscler Thromb. 2016 Aug 1;23(8):960-75. doi: 10.5551/jat.34462. Epub 2016 Feb 10. J Atheroscler Thromb. 2016. PMID: 26860885 Free PMC article. - Indoxyl glucuronide, a protein-bound uremic toxin, inhibits hypoxia-inducible factor‒dependent erythropoietin expression through activation of aryl hydrocarbon receptor.
Asai H, Hirata J, Watanabe-Akanuma M. Asai H, et al. Biochem Biophys Res Commun. 2018 Oct 2;504(2):538-544. doi: 10.1016/j.bbrc.2018.09.018. Epub 2018 Sep 8. Biochem Biophys Res Commun. 2018. PMID: 30205954 - Aryl Hydrocarbon Receptor Activation in Chronic Kidney Disease: Role of Uremic Toxins.
Brito JS, Borges NA, Esgalhado M, Magliano DC, Soulage CO, Mafra D. Brito JS, et al. Nephron. 2017;137(1):1-7. doi: 10.1159/000476074. Epub 2017 May 11. Nephron. 2017. PMID: 28490014 Review. - Targeting protein-bound uremic toxins in chronic kidney disease.
Niwa T. Niwa T. Expert Opin Ther Targets. 2013 Nov;17(11):1287-301. doi: 10.1517/14728222.2013.829456. Epub 2013 Aug 13. Expert Opin Ther Targets. 2013. PMID: 23941498 Review.
Cited by
- The Potential Influence of Uremic Toxins on the Homeostasis of Bones and Muscles in Chronic Kidney Disease.
Hung KC, Yao WC, Liu YL, Yang HJ, Liao MT, Chong K, Peng CH, Lu KC. Hung KC, et al. Biomedicines. 2023 Jul 24;11(7):2076. doi: 10.3390/biomedicines11072076. Biomedicines. 2023. PMID: 37509715 Free PMC article. Review. - The antifibrotic and anti-inflammatory effects of FZHY prescription on the kidney in rats after unilateral ureteral obstruction.
Chen Z, Wu S, Zeng Y, Li X, Wang M, Chen Z, Chen M. Chen Z, et al. Acta Cir Bras. 2023 Jan 6;37(10):e371003. doi: 10.1590/acb371003. eCollection 2023. Acta Cir Bras. 2023. PMID: 36629622 Free PMC article. - Indoxyl Sulfate Alters the Humoral Response of the ChAdOx1 COVID-19 Vaccine in Hemodialysis Patients.
Hou YC, Wu CL, Lu KC, Kuo KL. Hou YC, et al. Vaccines (Basel). 2022 Aug 24;10(9):1378. doi: 10.3390/vaccines10091378. Vaccines (Basel). 2022. PMID: 36146454 Free PMC article. - Chronic Kidney Failure Provokes the Enrichment of Terminally Differentiated CD8+ T Cells, Impairing Cytotoxic Mechanisms After Kidney Transplantation.
Leonhard J, Schaier M, Kälble F, Eckstein V, Zeier M, Steinborn A. Leonhard J, et al. Front Immunol. 2022 May 3;13:752570. doi: 10.3389/fimmu.2022.752570. eCollection 2022. Front Immunol. 2022. PMID: 35592311 Free PMC article. - Indoxyl Sulfate Elevated Lnc-SLC15A1-1 Upregulating CXCL10/CXCL8 Expression in High-Glucose Endothelial Cells by Sponging MicroRNAs.
Huang YC, Tsai TC, Chang CH, Chang KT, Ko PH, Lai LC. Huang YC, et al. Toxins (Basel). 2021 Dec 7;13(12):873. doi: 10.3390/toxins13120873. Toxins (Basel). 2021. PMID: 34941711 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous