Phage G Structure at 6.1 Å Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus - PubMed (original) (raw)
Phage G Structure at 6.1 Å Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus
Brenda González et al. J Mol Biol. 2020.
Abstract
Phage G has the largest capsid and genome of any known propagated phage. Many aspects of its structure, assembly, and replication have not been elucidated. Herein, we present the dsDNA-packed and empty phage G capsid at 6.1 and 9 Å resolution, respectively, using cryo-EM for structure determination and mass spectrometry for protein identification. The major capsid protein, gp27, is identified and found to share the HK97-fold universally conserved in all previously solved dsDNA phages. Trimers of the decoration protein, gp26, sit on the 3-fold axes and are thought to enhance the interactions of the hexameric capsomeres of gp27, for other phages encoding decoration proteins. Phage G's decoration protein is longer than what has been reported in other phages, and we suspect the extra interaction surface area helps stabilize the capsid. We identified several additional capsid proteins, including a candidate for the prohead protease responsible for processing gp27. Furthermore, cryo-EM reveals a range of partially full, condensed DNA densities that appear to have no contact with capsid shell. Three analyses confirm that the phage G host is a Lysinibacillus, and not Bacillus megaterium: identity of host proteins in our mass spectrometry analyses, genome sequence of the phage G host, and host range of phage G.
Keywords: DNA condensates; DNA packaging; Lysinibacillus; decoration proteins; phage G.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
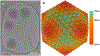
Figure 1:. Phage G capsid cryo-EM structure.
A) A representative micrograph shows particles with various capsid states. B) Phage G dsDNA-full capsid reconstruction at 6.1 Å resolution with 2,564 particles. The structure displayed is surface colored radially using the color bar on the right.
Figure 2:. Phage G major capsid protein hexamer and decoration protein trimer arrangement.
A) Phage G’s major capsid protein, gp27, and its decoration protein, gp26 arrangement. The structure shows gp27 hexamers and gp26 trimers positioned at the 3-fold axes around the hexamers. B) An overview of phage G’s gp27 homology model. All 3 domains of the structure are consistent with the domains described in HK97’s major capsid protein including the A domain, E loop, P domain. C) The phage G decoration, gp26, protein oligomerizes into trimers. The first 15 amino acids were omitted in modeling because of their flexibility. Arrows indicate the direction the N-terminus extends in the capsid density contacting neighboring gp27 subunits.
Figure 3:. Lack of concentric dsDNA rings in the phage G DNA-full structure.
A) Middle volume slice of phage G’s DNA-full capsid. B) Middle volume slice of phage G’s empty capsid. C) Radial distance plots were generated for the dsDNA-full (blue) and empty (orange) phage G structures.
Figure 4:. Multiple DNA states in phage G capsids seen in cryo-EM micrographs.
Multiple states of DNA packaged inside phage G capsid in our cryo-EM micrographs were categorized into 4 classes: full, empty, partial, and ruptured. Representative particles for each category are shown.
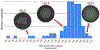
Figure 5:. Histogram of partial DNA diameter.
We further analyzed the group of 84 particles categorized as having partially full, compacted DNA density, by plotting a histogram of their DNA density width measured in the shortest dimension. The measured DNA density diameter values within the group of heterogeneous DNA density phage G capsids ranged from 495 Å to 1606 Å. Some particles appear to have toroid-like organizations from the side and top views as shown in the left two insets in this figure.
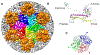
Figure 6:. Identification of major Phage G head proteins by mass spectrometry.
(A) Phage G genome region encoding the major head morphogenesis genes, including the terminase, portal, candidate prohead protease, major capsid protein (MCP) and head decoration protein (Dec). Genes whose products were identified by mass spectrometry are shaded blue. (B) SDS-PAGE gel of sucrose gradient purified Phage G virions. Individual gel slices that were subjected to mass spectrometric analyses are indicated. The mass spectral protein sequence coverage of the major capsid protein gp27 from different gel slices are shown in (C) gel slice 4, (D) gel slice 3, and (E) gel slice 2. Red arrow indicates cleavage by the prohead protease, gp19. The mass spectral protein sequence coverage of the head decoration protein gp26 from gel slice 2 is shown in (F). Amino-acids matched to a mass spectrum are shaded in yellow. Amino-acids marked in green potentially have a post-translational modification (e.g., phosphorylation).
Similar articles
- Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Bayfield OW, Klimuk E, Winkler DC, Hesketh EL, Chechik M, Cheng N, Dykeman EC, Minakhin L, Ranson NA, Severinov K, Steven AC, Antson AA. Bayfield OW, et al. Proc Natl Acad Sci U S A. 2019 Feb 26;116(9):3556-3561. doi: 10.1073/pnas.1813204116. Epub 2019 Feb 8. Proc Natl Acad Sci U S A. 2019. PMID: 30737287 Free PMC article. - Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo.
Fang PA, Wright ET, Weintraub ST, Hakala K, Wu W, Serwer P, Jiang W. Fang PA, et al. J Mol Biol. 2008 Dec 31;384(5):1384-99. doi: 10.1016/j.jmb.2008.10.012. Epub 2008 Oct 14. J Mol Biol. 2008. PMID: 18952096 Free PMC article. - Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure.
Stone NP, Demo G, Agnello E, Kelch BA. Stone NP, et al. Nat Commun. 2019 Oct 2;10(1):4471. doi: 10.1038/s41467-019-12341-z. Nat Commun. 2019. PMID: 31578335 Free PMC article. - Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins.
Dedeo CL, Teschke CM, Alexandrescu AT. Dedeo CL, et al. Viruses. 2020 Oct 14;12(10):1163. doi: 10.3390/v12101163. Viruses. 2020. PMID: 33066635 Free PMC article. Review. - Condensed genome structure.
Black LW, Thomas JA. Black LW, et al. Adv Exp Med Biol. 2012;726:469-87. doi: 10.1007/978-1-4614-0980-9_21. Adv Exp Med Biol. 2012. PMID: 22297527 Free PMC article. Review.
Cited by
- A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Podgorski JM, Freeman K, Gosselin S, Huet A, Conway JF, Bird M, Grecco J, Patel S, Jacobs-Sera D, Hatfull G, Gogarten JP, Ravantti J, White SJ. Podgorski JM, et al. Structure. 2023 Mar 2;31(3):282-294.e5. doi: 10.1016/j.str.2022.12.012. Epub 2023 Jan 16. Structure. 2023. PMID: 36649709 Free PMC article. - Characterization of Diverse Anelloviruses, Cressdnaviruses, and Bacteriophages in the Human Oral DNA Virome from North Carolina (USA).
Paietta EN, Kraberger S, Custer JM, Vargas KL, Espy C, Ehmke E, Yoder AD, Varsani A. Paietta EN, et al. Viruses. 2023 Aug 26;15(9):1821. doi: 10.3390/v15091821. Viruses. 2023. PMID: 37766228 Free PMC article. - Soil Giant Phage: Genome and Biological Characteristics of Sinorhizobium Jumbo Phage.
Kozlova AP, Muntyan VS, Vladimirova ME, Saksaganskaia AS, Kabilov MR, Gorbunova MK, Gorshkov AN, Grudinin MP, Simarov BV, Roumiantseva ML. Kozlova AP, et al. Int J Mol Sci. 2024 Jul 5;25(13):7388. doi: 10.3390/ijms25137388. Int J Mol Sci. 2024. PMID: 39000497 Free PMC article. - Structural Studies of the Phage G Tail Demonstrate an Atypical Tail Contraction.
González B, Li D, Li K, Wright ET, Hardies SC, Thomas JA, Serwer P, Jiang W. González B, et al. Viruses. 2021 Oct 18;13(10):2094. doi: 10.3390/v13102094. Viruses. 2021. PMID: 34696524 Free PMC article. - Viral Complexity.
Aylward FO, Moniruzzaman M. Aylward FO, et al. Biomolecules. 2022 Jul 30;12(8):1061. doi: 10.3390/biom12081061. Biomolecules. 2022. PMID: 36008955 Free PMC article. Review.
References
- Donelli G, Isolation of a bacteriophage of exceptional dimensions active in B megatherium, Atti Della Academia Nazionalez Dei Lincei Rendiconti-Classe Di Scienze, Fisichie, Matematiche, & Naturali. 44 (1968) 95.
- Donelli G, Griso G, Paoletti L, Rebessi S, Capsomeric Arrangement in the Bacteriophage G Head, in: Sixth Eur. Reg. Conf. Electron Microsc.(Jerusalem), 1976: pp. 502–503.
- Donelli G, Dore E, Frontali C, Grandolfo ME, Structure and physico-chemical properties of bacteriophage G: III. A homogeneous DNA of molecular weight 5 × 108, J. Mol. Biol 94 (1975) 555–565. - PubMed
- Ageno M, Donelli G, Guglielmi F, Structure and physico-chemical properties of bacteriophage G. II, The shape and symmetry of the capsid, Micron. 4 (1973) 376–403.
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM123055/GM/NIGMS NIH HHS/United States
- S10 RR025111/RR/NCRR NIH HHS/United States
- T32 GM132024/GM/NIGMS NIH HHS/United States
- UA5 GM126533/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources