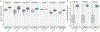PICRUSt2 for prediction of metagenome functions - PubMed (original) (raw)
PICRUSt2 for prediction of metagenome functions
Gavin M Douglas et al. Nat Biotechnol. 2020 Jun.
No abstract available
Figures
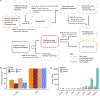
Figure 1:. PICRUSt2 algorithm.
(a) The PICRUSt2 method consists of phylogenetic placement, hidden-state-prediction and sample-wise gene and pathway abundance tabulation. ASV sequences and abundances are taken as input, and gene family and pathway abundances are output. All necessary reference tree and trait databases for the default workflow are included in the PICRUSt2 implementation. (b) The default PICRUSt1 pipeline restricted predictions to reference operational taxonomic units (Ref. OTUs) in the Greengenes database. This requirement resulted in the exclusion of many study sequences across four representative 16S rRNA gene sequencing datasets. PICRUSt2 relaxes this requirement and is agnostic to whether the input sequences are within a reference or not, which results in almost all of the input amplicon sequence variants (ASVs) being retained in the final output. (c) An increase in the taxonomic diversity in the default PICRUSt2 database is observed compared to PICRUSt1.
Figure 2:. PICRUSt2 performance characteristics.
Validation of PICRUSt2 KEGG ortholog (KO) predictions comparing metagenome prediction performance against gold-standard shotgun metagenomic sequencing (MGS). (a) Boxplots of Spearman correlation coefficients observed in stool samples from Cameroonian individuals (n=57), the human microbiome project (HMP, n=137), stool samples from Indian individuals (n=91), non-human primate stool samples (n=77), mammalian stool (n=8), ocean water (n=6), and blueberry soil (n=22) datasets. The significance of paired-sample, two-tailed Wilcoxon tests is indicated above each tested grouping (*, **, and ns correspond to P < 0.05, P < 0.001, and not significant respectively). (b) Comparison of significantly differentially abundant KOs between predicted metagenomes and MGS. Precision, recall, and F1 score are reported for each category compared to the MGS data. Precision corresponds to the proportion of significant KOs for that category also significant in the MGS data. Recall corresponds to the proportion of significant KOs in the MGS data also significant for that category. The F1 score is the harmonic mean of these metrics. The subsets of the four datasets compared are indicated above each panel (the Cameroonian parasite is Entamoeba). Wilcoxon tests were performed on the KO relative abundances after normalizing by the median number of universal single-copy genes per sample. Significance was defined at a false discovery rate < 0.05. The “Shuffled ASVs” category corresponds to PICRUSt2 predictions with ASV labels shuffled per dataset. The “Alt. MGS” category corresponds to an alternative MGS processing pipeline with reads aligned to the KEGG database rather than the default HUMAnN2 pipeline.
Figure 3:. PICRUSt2 accurately predicts MetaCyc pathways and phenotypes for characterizing overall environments.
(a) Spearman correlation coefficients between PICRUSt2 predicted pathway abundances and gold-standard metagenomic sequencing (MGS). Results are shown for each validation dataset: stool from Cameroonian individuals, The Human Microbiome Project (HMP), stool from Indian individuals, mammalian stool, ocean water, non-human primate stool, and blueberry soil. These results are limited to the 575 pathways that could potentially be identified by PICRUSt2 and HUMAnN2. (b) Performance of binary phenotype predictions based on three metrics: F1 score, precision, and recall. Each point corresponds to one of the 41 phenotypes tested. Predictions assessed here are based on holding out each genome individually, predicting the phenotypes for that holdout genome, and comparing the predicted and observed values. The null distribution in this case is based on randomizing the phenotypes across the reference genomes and comparing to the actual values, which results in the same output for all three metrics. The P-values of paired-sample, two-tailed Wilcoxon tests is indicated above each tested grouping (* and ** correspond to P < 0.05 and P < 0.001, respectively). Note that in panel a the y-axis is truncated below 0.5 rather than 0 to better visualize small differences between categories. The sample sizes in panel a are 57 (Cameroonian), 137 (HMP), 91 (Indian), 8 (mammal), 6 (ocean), 77 (primate), and 22 (soil).
References
Publication types
MeSH terms
Substances
Grants and funding
- F30 AA026527/AA/NIAAA NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R24 DK110499/DK/NIDDK NIH HHS/United States
- U54 DK102557/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases