Optimisation and standardisation of a multiplex immunoassay of diverse Plasmodium falciparum antigens to assess changes in malaria transmission using sero-epidemiology - PubMed (original) (raw)
doi: 10.12688/wellcomeopenres.14950.2. eCollection 2019.
Tom Hall 1, Isaac Ssewanyana 1 2, Tate Oulton 1, Catriona Patterson 1, Hristina Vasileva 1, Susheel Singh 3 4, Muna Affara 5, Julia Mwesigwa 6, Simon Correa 6, Mamadou Bah 6, Umberto D'Alessandro 6, Nuno Sepúlveda 1, Chris Drakeley 1, Kevin K A Tetteh 1
Affiliations
- PMID: 32518839
- PMCID: PMC7255915
- DOI: 10.12688/wellcomeopenres.14950.2
Optimisation and standardisation of a multiplex immunoassay of diverse Plasmodium falciparum antigens to assess changes in malaria transmission using sero-epidemiology
Lindsey Wu et al. Wellcome Open Res. 2020.
Abstract
Background: Antibody responses have been used to characterise transmission and exposure history in malaria-endemic settings for over a decade. Such studies have typically been conducted on well-standardised enzyme-linked immunosorbent assays (ELISAs). However, recently developed quantitative suspension array technologies (qSAT) are now capable of high-throughput and multiplexed screening of up to hundreds of analytes at a time. This study presents a customised protocol for the Luminex MAGPIX © qSAT using a diverse set of malaria antigens. The aim is to develop a standardised assay for routine serological surveillance that is implementable across laboratories and epidemiological settings. Methods: A panel of eight Plasmodium falciparum recombinant antigens, associated with long- and short-lived antibody responses, was designed for the Luminex MAGPIX © platform. The assay was optimised for key steps in the protocol: antigen-bead coupling concentration, buffer composition, serum sample dilution, and bead storage conditions. Quality control procedures and data normalisation methods were developed to address high-throughput assay processing. Antigen-specific limits of quantification (LOQs) were also estimated using both in-house and WHO reference serum as positive controls. Results: Antigen-specific bead coupling was optimised across five serum dilutions and two positive controls, resulting in concentrations operational within stable analytical ranges. Coupled beads were stable after storage at room temperature (22⁰C) for up to eight weeks. High sensitivity and specificity for distinguishing positive and negative controls at serum sample dilutions of 1:500 (AUC 0.94 95%CI 0.91-0.96) and 1:1000 (AUC 0.96 95%CI 0.94-0.98) were observed. LOQs were also successfully estimated for all analytes but varied by antigen and positive control. Conclusions: This study demonstrates that developing a standardised malaria-specific qSAT protocol for a diverse set of antigens is achievable, though further optimisations may be required. Quality control and data standardisation methods may also be useful for future analysis of large sero-epidemiological surveys.
Keywords: Luminex; MAGPIX; Plasmodium; antibodies; malaria; serology; serum.
Copyright: © 2020 Wu L et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
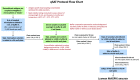
Figure 1.. Scheme describing the qSAT assay protocol.
Assay conditions tested for optimisation indicated in green boxes and red text.
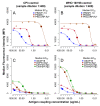
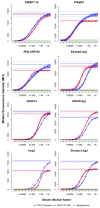
Figure 2.. Titration of antigen concentration for microsphere coupling using two positive controls at 1:400 serum dilution.
Antigens with maximum MFI values between 20,000 and 50,000 (_Pf_MSP119,_Pf_AMA1 and_Pf_GLURP.R2) shown in (A) and (B) and between 1,000 and 20,000 (Etramp5.Ag1, GEXP18, HSP40.Ag1, Etramp4.Ag2, Hyp2) in (C) and (D). Coupled microspheres were tested on two positive controls: CP3 (left) and WHO reference 10/198 (right). Optimal antigen coupling concentration (median EC50 across all sample dilutions and positive controls) are indicated as solid filled circles. *For_Pf_GLURP.R2, the two highest antigen concentrations (shown as triangles) were not used to fit standard curves to exclude the influence of prozone effect.
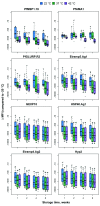
Figure 3.. Bead stability by temperature, storage time and dilution factor.
Difference in the median fluorescence intensity (∆MFI), of antigen-coupled beads stored at 22°C, 37°C and 42°C (compared to reference storage temperature of -20°C) after 1, 2, 4 and 8 weeks, and tested at six different positive control sample dilutions. Boxplots are based on data across all serum sample dilutions with median and interquartile range shown at each time point.
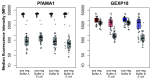
Figure 4.. Median fluorescence intensity (MFI) for _Pf_AMA1 and GEXP19 of positive and negative samples for each buffer composition.
Buffer compositions tested include buffer A (red), buffer A with_E. coli_ lysate (pink), buffer B (blue), and buffer B with_E. coli_ lysate (light blue). MFI for positive samples shown in colour (left) and corresponding negatives samples in grey (right) for each buffer composition.
Figure 5.. Limits of quantification.
Median fluorescence intensity (MFI) values are shown for pooled Tanzanian hyper-immune serum CP3 (blue) and WHO reference serum (red) in a 12-point serial dilution. Horizontal lines represent the mean and 95% confidence intervals of the upper and lower asymptotes of the sigmoidal curve for CP3 (blue), WHO (red), and the mean background MFI in green.
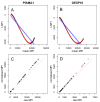
Figure 6.. Loess normalisation for _Pf_AMA1 and GEXP18.
Loess normalisation of antigens_Pf_AMA1 and GEXP18 is illustrated for one example plate based on cross-sectional samples from The Gambia. PanelsA andB show the loess fit (red) and linear fit (blue) of ∆ MFI on the y-axis (the difference in unadjusted MFI of the standard curve of a single plate and the mean MFI of all standard curves from ten reference plates) and mean MFI on the x-axis. PanelsC andD show the raw MFI values of individual samples on the x-axis versus normalised MFI values on the y-axis, and the equality line is shown diagonally in red.
Similar articles
- Development of a new peptide-bead coupling method for an all peptide-based Luminex multiplexing assay for detection of Plasmodium falciparum antibody responses.
Wakeman BS, Shakamuri P, McDonald MA, Weinberg J, Svoboda P, Murphy MK, Kariuki S, Mace K, Elder E, Rivera H, Qvarnstrom Y, Pohl J, Shi YP. Wakeman BS, et al. J Immunol Methods. 2021 Dec;499:113148. doi: 10.1016/j.jim.2021.113148. Epub 2021 Sep 21. J Immunol Methods. 2021. PMID: 34560073 - Optimization of incubation conditions of Plasmodium falciparum antibody multiplex assays to measure IgG, IgG1-4, IgM and IgE using standard and customized reference pools for sero-epidemiological and vaccine studies.
Ubillos I, Jiménez A, Vidal M, Bowyer PW, Gaur D, Dutta S, Gamain B, Coppel R, Chauhan V, Lanar D, Chitnis C, Angov E, Beeson J, Cavanagh D, Campo JJ, Aguilar R, Dobaño C. Ubillos I, et al. Malar J. 2018 Jun 1;17(1):219. doi: 10.1186/s12936-018-2369-3. Malar J. 2018. PMID: 29859096 Free PMC article. - Development of a high-throughput flexible quantitative suspension array assay for IgG against multiple Plasmodium falciparum antigens.
Ubillos I, Campo JJ, Jiménez A, Dobaño C. Ubillos I, et al. Malar J. 2018 May 29;17(1):216. doi: 10.1186/s12936-018-2365-7. Malar J. 2018. PMID: 29843713 Free PMC article. - A systematic review on malaria sero-epidemiology studies in the Brazilian Amazon: insights into immunological markers for exposure and protection.
Folegatti PM, Siqueira AM, Monteiro WM, Lacerda MV, Drakeley CJ, Braga ÉM. Folegatti PM, et al. Malar J. 2017 Mar 7;16(1):107. doi: 10.1186/s12936-017-1762-7. Malar J. 2017. PMID: 28270152 Free PMC article. Review. - Research priorities for the development and implementation of serological tools for malaria surveillance.
Elliott SR, Fowkes FJ, Richards JS, Reiling L, Drew DR, Beeson JG. Elliott SR, et al. F1000Prime Rep. 2014 Nov 4;6:100. doi: 10.12703/P6-100. eCollection 2014. F1000Prime Rep. 2014. PMID: 25580254 Free PMC article. Review.
Cited by
- Higher gametocyte production and mosquito infectivity in chronic compared to incident Plasmodium falciparum infections.
Barry A, Bradley J, Stone W, Guelbeogo MW, Lanke K, Ouedraogo A, Soulama I, Nébié I, Serme SS, Grignard L, Patterson C, Wu L, Briggs JJ, Janson O, Awandu SS, Ouedraogo M, Tarama CW, Kargougou D, Zongo S, Sirima SB, Marti M, Drakeley C, Tiono AB, Bousema T. Barry A, et al. Nat Commun. 2021 Apr 26;12(1):2443. doi: 10.1038/s41467-021-22573-7. Nat Commun. 2021. PMID: 33903595 Free PMC article. - Characterizing the spatial distribution of multiple malaria diagnostic endpoints in a low-transmission setting in Lao PDR.
Byrne I, Cramer E, Nelli L, Rerolle F, Wu L, Patterson C, Rosado J, Dumont E, Tetteh KKA, Dantzer E, Hongvanthong B, Fornace KM, Stresman G, Lover A, Bennett A, Drakeley C. Byrne I, et al. Front Med (Lausanne). 2022 Aug 18;9:929366. doi: 10.3389/fmed.2022.929366. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36059850 Free PMC article. - Current malaria infection, previous malaria exposure, and clinical profiles and outcomes of COVID-19 in a setting of high malaria transmission: an exploratory cohort study in Uganda.
Achan J, Serwanga A, Wanzira H, Kyagulanyi T, Nuwa A, Magumba G, Kusasira S, Sewanyana I, Tetteh K, Drakeley C, Nakwagala F, Aanyu H, Opigo J, Hamade P, Marasciulo M, Baterana B, Tibenderana JK. Achan J, et al. Lancet Microbe. 2022 Jan;3(1):e62-e71. doi: 10.1016/S2666-5247(21)00240-8. Epub 2021 Oct 25. Lancet Microbe. 2022. PMID: 34723228 Free PMC article. - Long-Term SARS-CoV-2-Specific Immune and Inflammatory Responses Across a Clinically Diverse Cohort of Individuals Recovering from COVID-19.
Peluso MJ, Deitchman AN, Torres L, Iyer NS, Nixon CC, Munter SE, Donatelli J, Thanh C, Takahashi S, Hakim J, Turcios K, Janson O, Hoh R, Tai V, Hernandez Y, Fehrman E, Spinelli MA, Gandhi M, Trinh L, Wrin T, Petropoulos CJ, Aweeka FT, Rodriguez-Barraquer I, Kelly JD, Martin JN, Deeks SG, Greenhouse B, Rutishauser RL, Henrich TJ. Peluso MJ, et al. medRxiv [Preprint]. 2021 Mar 1:2021.02.26.21252308. doi: 10.1101/2021.02.26.21252308. medRxiv. 2021. PMID: 33688685 Free PMC article. Updated. Preprint. - Malaria seroepidemiology in very low transmission settings in the Peruvian Amazon.
Fernandez-Camacho B, Peña-Calero B, Guillermo-Roman M, Ruiz-Cabrejos J, Barboza JL, Bartolini-Arana L, Barja-Ingaruca A, Rodriguez-Ferrucci H, Soto-Calle VE, Nelli L, Byrne I, Hill M, Dumont E, Grignard L, Tetteh K, Wu L, Llanos-Cuentas A, Drakeley C, Stresman G, Carrasco-Escobar G. Fernandez-Camacho B, et al. Sci Rep. 2024 Feb 2;14(1):2806. doi: 10.1038/s41598-024-52239-5. Sci Rep. 2024. PMID: 38307878 Free PMC article.
References
LinkOut - more resources
Full Text Sources