Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers - PubMed (original) (raw)
. 2020 May 29;6(22):eaaz5344.
doi: 10.1126/sciadv.aaz5344. eCollection 2020 May.
Choongwon Jeong 2 3, Stephan Schiffels 2, İşil Küçükkalıpçı 2, Marie-Hélène Pemonge 1, Adam Benjamin Rohrlach 2 4, Kurt W Alt 5 6, Didier Binder 7, Susanne Friederich 8, Emmanuel Ghesquière 9 10, Detlef Gronenborn 11, Luc Laporte 10, Philippe Lefranc 12 13, Harald Meller 8, Hélène Réveillas 1 14, Eva Rosenstock 15 16, Stéphane Rottier 1, Chris Scarre 17, Ludovic Soler 1 18, Joachim Wahl 19 20, Johannes Krause 2, Marie-France Deguilloux 1, Wolfgang Haak 2
Affiliations
- PMID: 32523989
- PMCID: PMC7259947
- DOI: 10.1126/sciadv.aaz5344
Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers
Maïté Rivollat et al. Sci Adv. 2020.
Abstract
Starting from 12,000 years ago in the Middle East, the Neolithic lifestyle spread across Europe via separate continental and Mediterranean routes. Genomes from early European farmers have shown a clear Near Eastern/Anatolian genetic affinity with limited contribution from hunter-gatherers. However, no genomic data are available from modern-day France, where both routes converged, as evidenced by a mosaic cultural pattern. Here, we present genome-wide data from 101 individuals from 12 sites covering today's France and Germany from the Mesolithic (N = 3) to the Neolithic (N = 98) (7000-3000 BCE). Using the genetic substructure observed in European hunter-gatherers, we characterize diverse patterns of admixture in different regions, consistent with both routes of expansion. Early western European farmers show a higher proportion of distinctly western hunter-gatherer ancestry compared to central/southeastern farmers. Our data highlight the complexity of the biological interactions during the Neolithic expansion by revealing major regional variations.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures
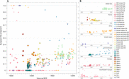
Fig. 1. Spatial, temporal, and genetic structure of individuals in this study.
(A) Geographic map showing sample locations. (B) Chronological timeline: Maximal chronological range according to calibrated radiocarbon date ranges (2-sigma) for each site/individual. (C) Principal components analysis (zoom in). Published ancient (no outlined symbols) and newly reported (black outlined symbols) individuals projected onto 777 present-day west Eurasians (gray circles).
Fig. 2. HG ancestry proportions over time.
(A) Overall timeline: Results of qpAdm (MODEL A) modeling of European_HG (represented by Loschbour, La Braña, and KO1; y axis) and Anatolia_Neolithic ancestry for each individual with a direct radiocarbon date ranging between 6200 and 2800 BCE (x axis). See table S10 for further details. (B) Regional timelines: Plots of the data shown in (A), separated by geographic location, to show regional signals.
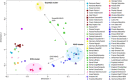
Fig. 3. Multidimensional scaling plot HG individuals.
Multidimensional scaling plot of genetic distances based on _f_-statistics of the form f3(Mbuti; test, test) between Eurasian HG individuals. Newly reported individuals have a black outline.
Fig. 4. Maps of variable sources of HG and Anatolian Neolithic ancestries through time.
(A) Proportion of distal sources of Villabruna, EHG, Goyet_Q2, and Anatolia_Neolithic of post-LGM HG individuals (14000–4000 cal BCE) estimated according to MODEL B (table S12). The following five panels (B to F) show Neolithic farmer groups (6000–3500 cal BCE) modeled with the proximal sources Anatolia_Neolithic, KO1, and Loschbour according to MODEL C, in time increments of 500 years each. Transparent colors indicate individuals or groups not sufficiently supported by the models (P < 0.05). Note that not only N22 from the Polish BKG, TGM009, and Blätterhöhle, which carry a substantial proportion of HG ancestry, but also Anatolian_Neolithic ancestry have been modeled with MODEL B and were added to (E) and (F), respectively. See table S14 for model details.
Fig. 5. Admixture dating per group.
Admixture date estimation according to DATES software, obtained in generations. Radiocarbon date intervals (given with 2-sigma) for each site are black lines; blue diamonds are the estimated admixture date (SE = 1). Admixture is estimated with two sources: European_HG and Anatolia_Neolithic. Number of years calculated on the basis of 28 years for one generation. Admixture time calculated according to the oldest date of the radiocarbon interval for each group (table S17 and text S10).
Similar articles
- Heterogeneous Hunter-Gatherer and Steppe-Related Ancestries in Late Neolithic and Bell Beaker Genomes from Present-Day France.
Seguin-Orlando A, Donat R, Der Sarkissian C, Southon J, Thèves C, Manen C, Tchérémissinoff Y, Crubézy E, Shapiro B, Deleuze JF, Dalén L, Guilaine J, Orlando L. Seguin-Orlando A, et al. Curr Biol. 2021 Mar 8;31(5):1072-1083.e10. doi: 10.1016/j.cub.2020.12.015. Curr Biol. 2021. PMID: 33434506 - Paleogenomic Evidence for Multi-generational Mixing between Neolithic Farmers and Mesolithic Hunter-Gatherers in the Lower Danube Basin.
González-Fortes G, Jones ER, Lightfoot E, Bonsall C, Lazar C, Grandal-d'Anglade A, Garralda MD, Drak L, Siska V, Simalcsik A, Boroneanţ A, Vidal Romaní JR, Vaqueiro Rodríguez M, Arias P, Pinhasi R, Manica A, Hofreiter M. González-Fortes G, et al. Curr Biol. 2017 Jun 19;27(12):1801-1810.e10. doi: 10.1016/j.cub.2017.05.023. Epub 2017 May 25. Curr Biol. 2017. PMID: 28552360 Free PMC article. - Hunter-gatherer admixture facilitated natural selection in Neolithic European farmers.
Davy T, Ju D, Mathieson I, Skoglund P. Davy T, et al. Curr Biol. 2023 Apr 10;33(7):1365-1371.e3. doi: 10.1016/j.cub.2023.02.049. Epub 2023 Mar 23. Curr Biol. 2023. PMID: 36963383 Free PMC article. - A genomic Neolithic time transect of hunter-farmer admixture in central Poland.
Fernandes DM, Strapagiel D, Borówka P, Marciniak B, Żądzińska E, Sirak K, Siska V, Grygiel R, Carlsson J, Manica A, Lorkiewicz W, Pinhasi R. Fernandes DM, et al. Sci Rep. 2018 Oct 5;8(1):14879. doi: 10.1038/s41598-018-33067-w. Sci Rep. 2018. PMID: 30291256 Free PMC article. - The evolutionary history of human populations in Europe.
Lazaridis I. Lazaridis I. Curr Opin Genet Dev. 2018 Dec;53:21-27. doi: 10.1016/j.gde.2018.06.007. Epub 2018 Jun 28. Curr Opin Genet Dev. 2018. PMID: 29960127 Review.
Cited by
- The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes.
Mallick S, Micco A, Mah M, Ringbauer H, Lazaridis I, Olalde I, Patterson N, Reich D. Mallick S, et al. Sci Data. 2024 Feb 10;11(1):182. doi: 10.1038/s41597-024-03031-7. Sci Data. 2024. PMID: 38341426 Free PMC article. - Genetic Variability and Population Structure of the Tunisian Sicilo-Sarde Dairy Sheep Breed Inferred from Microsatellites Analysis.
Ben Sassi-Zaidy Y, Mohamed-Brahmi A, Nouairia G, Charfi-Cheikhrouha F, Djemali M, Cassandro M. Ben Sassi-Zaidy Y, et al. Genes (Basel). 2022 Feb 5;13(2):304. doi: 10.3390/genes13020304. Genes (Basel). 2022. PMID: 35205349 Free PMC article. - Neolithic genomic data from southern France showcase intensified interactions with hunter-gatherer communities.
Arzelier A, Rivollat M, De Belvalet H, Pemonge MH, Binder D, Convertini F, Duday H, Gandelin M, Guilaine J, Haak W, Deguilloux MF, Pruvost M. Arzelier A, et al. iScience. 2022 Oct 19;25(11):105387. doi: 10.1016/j.isci.2022.105387. eCollection 2022 Nov 18. iScience. 2022. PMID: 36405775 Free PMC article. - Population Genetics and Signatures of Selection in Early Neolithic European Farmers.
Childebayeva A, Rohrlach AB, Barquera R, Rivollat M, Aron F, Szolek A, Kohlbacher O, Nicklisch N, Alt KW, Gronenborn D, Meller H, Friederich S, Prüfer K, Deguilloux MF, Krause J, Haak W. Childebayeva A, et al. Mol Biol Evol. 2022 Jun 2;39(6):msac108. doi: 10.1093/molbev/msac108. Mol Biol Evol. 2022. PMID: 35578825 Free PMC article. - Interdisciplinary Analyses of Bronze Age Communities from Western Hungary Reveal Complex Population Histories.
Gerber D, Szeifert B, Székely O, Egyed B, Gyuris B, Giblin JI, Horváth A, Köhler K, Kulcsár G, Kustár Á, Major I, Molnár M, Palcsu L, Szeverényi V, Fábián S, Mende BG, Bondár M, Ari E, Kiss V, Szécsényi-Nagy A. Gerber D, et al. Mol Biol Evol. 2023 Sep 1;40(9):msad182. doi: 10.1093/molbev/msad182. Mol Biol Evol. 2023. PMID: 37562011 Free PMC article.
References
- D. Gronenborn, The persistence of hunting and gathering: Neolithic western temperate and Central Europe, in The Oxford Handbook of the Archaeology and Anthropology of Hunter-Gatherers, V. Cummings, P. Jordan, M. Zvelebil, Eds. (Oxford University Press, 2014), pp. 787–804.
- J. Guilaine, Aspects de la néolithisation en Méditerranée et en France, in The Widening Harvest. The Neolithic Transition in Europe: Looking Back, Looking Forward, A. Ammerman, P. Biagi, Eds. (Archaeological Institute of America, 2003), pp. 189–206.
- Bramanti B., Thomas M. G., Haak W., Unterlaender M., Jores P., Tambets K., Antanaitis-Jacobs I., Haidle M. N., Jankauskas R., Kind C.-J., Lueth F., Terberger T., Hiller J., Matsumura S., Forster P., Burger J., Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science 326, 137–140 (2009). - PubMed
- Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K., Fu Q., Mittnik A., Bánffy E., Economou C., Francken M., Friederich S., Garrido Pena R., Hallgren F., Khartanovich V., Khokhlov A., Kunst M., Kuznetsov P., Meller H., Mochalov O., Moiseyev V., Nicklisch N., Pichler S. L., Risch R., Rojo Guerra M. A., Roth C., Szécsényi-Nagy A., Wahl J., Meyer M., Krause J., Brown D., Anthony D., Cooper A., Alt K. W., Reich D., Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 552, 207–211 (2015). - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources