Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity - PubMed (original) (raw)
Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity
Thomas Dahlet et al. Nat Commun. 2020.
Abstract
Mouse embryos acquire global DNA methylation of their genome during implantation. However the exact roles of DNA methyltransferases (DNMTs) in embryos have not been studied comprehensively. Here we systematically analyze the consequences of genetic inactivation of Dnmt1, Dnmt3a and Dnmt3b on the methylome and transcriptome of mouse embryos. We find a strict division of function between DNMT1, responsible for maintenance methylation, and DNMT3A/B, solely responsible for methylation acquisition in development. By analyzing severely hypomethylated embryos, we uncover multiple functions of DNA methylation that is used as a mechanism of repression for a panel of genes including not only imprinted and germline genes, but also lineage-committed genes and 2-cell genes. DNA methylation also suppresses multiple retrotransposons and illegitimate transcripts from cryptic promoters in transposons and gene bodies. Our work provides a thorough analysis of the roles of DNA methyltransferases and the importance of DNA methylation for transcriptome integrity in mammalian embryos.
Conflict of interest statement
The authors declare no competing interests.
Figures
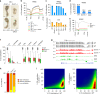
Fig. 1. Methylome profiling in Dnmt mutant embryos.
a Images of Dnmt1 _−/_− and DKO embryos dissected at E8.5 compared with littermate heterozygous controls (which are identical to WT). Black bars: 300 μm. b Average distribution of CG methylation over RefSeq genes and 10 kb flanking sequences calculated from the RRBS data in E8.5 embryos. Our previous data from single Dnmt3a −/− and Dnmt3b −/− embryos was included for comparison. TSS transcription start site, TTS transcription termination site. c Average methylation of CGs located outside of CpG islands (non-CGI, top) or within CpG islands (CGI, bottom) measured by RRBS in Dnmt mutant and control embryos (mean ± SEM, n = 3 independent embryos for all genotypes except n = 2 for Dnmt3a −/− and Dnmt3b −/−). d Bar plot representing the average CG methylation level measured by WGBS in WT, Dnmt1 −/− and DKO E8.5 embryos (mean of n = 2 independent embryos). e Metaplots of CG methylation levels in RefSeq genes and 10 kb flanking sequences calculated from the WGBS data in WT, Dnmt1 _−/_− and DKO E8.5 embryos. f Boxplots of CG methylation levels measured by WGBS in different genomic features in WT, Dnmt1 −/− and DKO E8.5 embryos. In the boxplots the line indicates the median, the box limits indicate the upper and lower quartiles and the whiskers extend to 1.5 IQR from the quartiles. g Example of genome browser view of WGBS methylation profiles in two independent replicates of WT, Dnmt1 _−/_− and DKO E8.5 embryos. Each track shows the percent methylation of individual CpGs between 0 and 100%. CpG islands (CGIs) depicted by green rectangles and RefSeq gene annotations are shown below the tracks. h Stacked bar graph representing the proportions of genomic windows (1 kb) with high (>50%), medium (10–50%), and low (<10%) CG methylation in WT, Dnmt1 _−/_− and DKO embryos. i Density scatter plots comparing WGBS methylation scores in 500 bp windows between WT, Dnmt1 −/− and DKO E8.5 embryos. The density of points increases from blue to dark red. In f, h, i, values are the average of n = 2 independent embryos. Source data are provided as a Source data file.
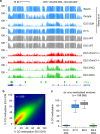
Fig. 2. Absence of de novo methylation in DKO embryos.
a Genome browser tracks of WGBS methylation profiles in gametes, early embryos, and E8.5 Dnmt mutant embryos, highlighting the strong similarity of methylation patterns between the ICM (Inner Cell Mass) and DKO embryos. CpG islands (CGIs) depicted by green rectangles and RefSeq gene annotations are shown below the tracks. b Density scatter plot of WGBS methylation scores in 1 kb windows in E8.5 DKO embryos compared with E3.5 ICM. The density of points increases from blue to dark red. c Boxplots of the CG methylation levels in ICM, postimplantation embryos, and E8.5 DKO embryos for all genomic windows de novo methylated at implantation, illustrating that de novo methylation is abolished in DKO embryos. In the boxplots the line indicates the median, the box limits indicate the upper and lower quartiles and the whiskers extend to 1.5 IQR from the quartiles.
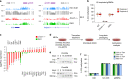
Fig. 3. Role of DNMTs in methylation imprints and maintenance methylation.
a Genome browser tracks of WGBS methylation profiles at the H19 and Impact imprinted loci in early embryos and Dnmt mutant embryos. One WGBS replicate is shown. The positions of the imprinted germline DMRs (gDMRs) are depicted by purple rectangles. b Boxplots of the CG methylation levels of n = 20 imprinted germline DMRs measured by WGBS in WT, Dnmt1 −/− and DKO embryos. In the boxplots the line indicates the median, the box limits indicate the upper and lower quartiles and the whiskers extend to 1.5 IQR from the quartiles. c Fold change of expression of imprinted genes in Dnmt1 −/− (red bars) and DKO (green bars) embryos (n = 3 embryos for Dnmt1 −/−; n = 6 embryos for DKO). **p < 0.01; ***p < 0.001 (adjusted p value calculated by DESeq2 using a Wald test corrected for multiple testing). d Experimental outline for investigating the maintenance function of DNMT3A/B in MEFs. Dnmt3a 2lox/2lox Dnmt3b 2lox/2lox immortalized MEFs expressing Cre-ERT2 were treated with Tamoxifen to generate conditional double knockout (cDKO) cells, and methylation was quantified by RRBS after long-term passaging. e Evaluation by PCR genotyping of the Cre-mediated recombination of the Dnmt3a and _Dnmt3b_-2lox alleles in cDKO fibroblasts. The number of days of Tamoxifen treatment is indicated above the gels. f CG methylation levels quantified by RRBS in cDKO fibroblasts. The graph shows the methylation levels in CpG islands (CGI), non-CGI regions, and imprinted gDMRs in cells treated with Tamoxifen (Tam) or not treated with Tamoxifen (no Tam) after 23 and 69 days of culture (mean ± SEM, n = 3 biological replicates), revealing no global hypomethylation in cDKO MEFs. Source data are provided as a Source data file.
Fig. 4. Transcriptome analysis of Dnmt mutant embryos.
a Principal component analysis of RNA-seq data. b Venn diagram comparing the lists of upregulated and downregulated genes in Dnmt1 −/− and DKO embryos. c Heatmap of the three groups of genes upregulated in DKO embryos classified by their promoter class (LCP low CpG promoter, ICP intermediate CpG promoter, HCP high CpG promoter) and promoter methylation in WT embryos (measured in −1000 to +500 bp from the TSS). Group 1: LCP; Group 2: ICP or HCP and promoter methylation < 30%; Group 3: ICP or HCP and promoter methylation ≥ 30%. Gene ontology (GO) terms enriched in each group are shown on the right. d Expression levels (FPKM) of germline genes in DKO and control embryos (mean ± SEM, n = 6 embryos). e RT-qPCR expression levels of Dazl and Asz1 in non-transfected MEFs (NT) and MEFs expressing dCas9-Suntag-TET1 with no gRNA or a gRNA targeting the Dazl or Asz1 promoter (mean ± SEM, n = 3 independent experiments). Cells treated with 0.5 μM 5-Aza-2′-deoxycytidine (5azadC) for 72 h were used as control. *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed unpaired t test). f Expression levels (FPKM) of lineage-committed genes in DKO, Dnmt1 _−/_− and control embryos (mean ± SEM, n = 6 embryos for control and DKO, n = 3 for Dnmt1 −/−). g RT-qPCR analysis of the expression of five lineage-committed genes in control and DKO embryos (mean ± SEM, n = 4 embryos). *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed unpaired t test). h Genome browser tracks of RNA-seq and WGBS at the hematopoietic-specific Bin2 gene in WT and DKO embryos and adult tissues–. For embryos, one replicate of RNA-seq and WGBS is shown. The Bin2 promoter (highlighted in yellow) is specifically hypomethylated in hematopoietic tissues (written in red). i Expression levels (FPKM) of 2C-specific genes in DKO, Dnmt1 _−/_− and control embryos (mean ± SEM, n = 6 embryos for control and DKO, n = 3 for Dnmt1 _−/_−). j RT-qPCR analysis of the expression of 2C-specific genes in control and DKO embryos (mean ± SEM, n = 5 embryos). The p values are indicated (two-tailed unpaired t test). Source data are provided as a Source data file.
Fig. 5. Dnmt mutant embryos show widespread transposon upregulation.
a DNA methylation measured by WGBS and fold change of expression of SINE, LINE, ERV1, ERVK, and EVRL retrotransposons in Dnmt1 _−/_− and DKO embryos. Methylation and expression was calculated on RepeatMasker annotations. b Percentage of significantly upregulated copies within each retrotransposon family in Dnmt1 _−/_− embryos. c Boxplots comparing the size of transposon copies upregulated in Dnmt1 _−/_− embryos (up) or not upregulated (not up) for several ERV families. ***p < 0.001 (Wilcoxon test). In the boxplots the line indicates the median, the box limits indicate the upper and lower quartiles and the whiskers extend to 1.5 IQR from the quartiles. IAPEz-int: n = 4835 (not up), n = 2484 (up), p = 0; MMERVK10C-int: n = 3193 (not up), n = 37 (up), p = 1.53e−14; MMERGLN-int: n = 834 (not up), n = 22 (up), p = 2.15e−12; ERVB4_1B-I_MM-int: n = 1347 (not up), n = 21 (up), p = 8.03e−12; MMETn-int: n = 936 (not up), n = 44 (up), p = 3.97e−14. d Fold change of expression of genes located close to upregulated ERVs (genes ERV up, n = 715) compared with all genes, genes located close to all ERVs (genes ERV all) and a random selection of 715 genes located close to ERVs (genes ERV rand). In the boxplots the line indicates the median, the dot indicates the mean, the box limits indicate the upper and lower quartiles and the whiskers extend to 1.5 IQR from the quartiles. ***p < 0.001 (Wilcoxon test). e Percentage of significantly upregulated genes in Dnmt1 _−/_− embryos for genes located close to upregulated ERVs and control genes. f Cyp2b23 expression is induced by an upstream cluster of ERVs, which initiates a long RNA that splices into the exon 2. The figure shows RNA-seq tracks in WT and Dnmt1 −/− embryos, along with splice junctions in one replicate of WT and Dnmt1 −/− embryo. ERVs annotated by RepeatMasker are displayed in yellow. g RNA-seq tracks in WT and Dnmt1 _−/_− embryos illustrating that the derepression of an intragenic IAPEz element leads to internal initiation of the Capn11 gene on the opposite strand. The RNA-seq signals from the top (above the line) and bottom (below the line) strands are shown. IAPs annotated by RepeatMasker are displayed in yellow.
Fig. 6. Transcripts initiate from cryptic intragenic promoters in Dnmt mutant embryos.
a Genome browser tracks of WGBS and RNA-seq profiles at the Mgl2 locus in WT and Dnmt mutant embryos. One WGBS replicates and three RNA-seq replicates are shown. RepeatMasker annotations are displayed in yellow below the tracks. b Boxplot of the ratio of RNA-seq read counts in downstream exons compared to the first exon in WT and DKO embryos for all expressed genes with at least 5 exons (n = 12,898). Exon n represents the last exon of the gene. ns: not significant; *p < 0.05; **p < 0.01; ***p < 0.001 (Wilcoxon test). c Metaplots representing the CpG density and the CpG methylation levels in 5 kb sequences flanking cryptic intragenic initiation sites or canonical RefSeq TSS. d Genome browser tracks of RNA-seq, DNAse-seq and NRF1 ChIP-seq profiles at the Mgl2 locus in WT and Dnmt triple knockout (TKO) ES cells. Two replicates are shown. e The genome browser tracks on the top display Mgl2 RNA-seq profiles upon NRF1 knockdown in TKO ES cells. The boxplot on the bottom shows the quantification of Mgl2 expression measured by RNA-seq (plotted as FPKM) upon mock and NRF1 knockdown in TKO ES cells (n = 3 replicates). In the boxplots the line indicates the median, the box limits indicate the upper and lower quartiles and the whiskers extend to 1.5 IQR from the quartiles.
Similar articles
- DNMT1, DNMT3A and DNMT3B proteins are differently expressed in mouse oocytes and early embryos.
Uysal F, Ozturk S, Akkoyunlu G. Uysal F, et al. J Mol Histol. 2017 Dec;48(5-6):417-426. doi: 10.1007/s10735-017-9739-y. Epub 2017 Oct 13. J Mol Histol. 2017. PMID: 29027601 - DNMT1 in Six2 Progenitor Cells Is Essential for Transposable Element Silencing and Kidney Development.
Li SY, Park J, Guan Y, Chung K, Shrestha R, Palmer MB, Susztak K. Li SY, et al. J Am Soc Nephrol. 2019 Apr;30(4):594-609. doi: 10.1681/ASN.2018070687. Epub 2019 Mar 8. J Am Soc Nephrol. 2019. PMID: 30850438 Free PMC article. - Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development.
Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Hirasawa R, et al. Genes Dev. 2008 Jun 15;22(12):1607-16. doi: 10.1101/gad.1667008. Genes Dev. 2008. PMID: 18559477 Free PMC article. - Role of Mammalian DNA Methyltransferases in Development.
Chen Z, Zhang Y. Chen Z, et al. Annu Rev Biochem. 2020 Jun 20;89:135-158. doi: 10.1146/annurev-biochem-103019-102815. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815535 Review. - DNA methyltransferases in hematological malignancies.
Hoang NM, Rui L. Hoang NM, et al. J Genet Genomics. 2020 Jul 20;47(7):361-372. doi: 10.1016/j.jgg.2020.04.006. Epub 2020 Jul 24. J Genet Genomics. 2020. PMID: 32994141 Free PMC article. Review.
Cited by
- ZIC2 and ZIC3 promote SWI/SNF recruitment to safeguard progression towards human primed pluripotency.
Hossain I, Priam P, Reynoso SC, Sahni S, Zhang XX, Côté L, Doumat J, Chik C, Fu T, Lessard JA, Pastor WA. Hossain I, et al. Nat Commun. 2024 Oct 2;15(1):8539. doi: 10.1038/s41467-024-52431-1. Nat Commun. 2024. PMID: 39358345 Free PMC article. - 3t-seq: automatic gene expression analysis of single-copy genes, transposable elements, and tRNAs from RNA-seq data.
Tabaro F, Boulard M. Tabaro F, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae467. doi: 10.1093/bib/bbae467. Brief Bioinform. 2024. PMID: 39322626 Free PMC article. - Unraveling the genetic and epigenetic landscape governing intramuscular fat deposition in rabbits: Insights and implications.
Ahamba IS, Mary-Cynthia Ikele C, Kimpe L, Goswami N, Wang H, Li Z, Ren Z, Dong X. Ahamba IS, et al. Food Chem (Oxf). 2024 Aug 30;9:100222. doi: 10.1016/j.fochms.2024.100222. eCollection 2024 Dec 30. Food Chem (Oxf). 2024. PMID: 39290671 Free PMC article. Review. - The proteomic landscape and temporal dynamics of mammalian gastruloid development.
Garge RK, Lynch V, Fields R, Casadei S, Best S, Stone J, Snyder M, McGann CD, Shendure J, Starita LM, Hamazaki N, Schweppe DK. Garge RK, et al. bioRxiv [Preprint]. 2024 Sep 7:2024.09.05.609098. doi: 10.1101/2024.09.05.609098. bioRxiv. 2024. PMID: 39282277 Free PMC article. Preprint. - Kick-starting the zygotic genome: licensors, specifiers, and beyond.
Zou Z, Wang Q, Wu X, Schultz RM, Xie W. Zou Z, et al. EMBO Rep. 2024 Oct;25(10):4113-4130. doi: 10.1038/s44319-024-00223-5. Epub 2024 Aug 19. EMBO Rep. 2024. PMID: 39160344 Free PMC article. Review.
References
- Smith ZD, et al. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. - PubMed
- Bourc’his D, et al. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. - PubMed
- Barau J, et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016;354:909–912. - PubMed
- Okano M, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
- Li E, et al. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials