A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2 - PubMed (original) (raw)
. 2020 Aug 7;369(6504):650-655.
doi: 10.1126/science.abc6952. Epub 2020 Jun 22.
Renhong Yan # 2, Jun Zhang # 1, Guanying Zhang 1, Yuanyuan Zhang 2, Meng Hao 1, Zhe Zhang 1, Pengfei Fan 1, Yunzhu Dong 1, Yilong Yang 1, Zhengshan Chen 1, Yingying Guo 2, Jinlong Zhang 1, Yaning Li 3, Xiaohong Song 1, Yi Chen 1, Lu Xia 2, Ling Fu 1, Lihua Hou 1, Junjie Xu 1, Changming Yu 1, Jianmin Li 4, Qiang Zhou 5, Wei Chen 4
Affiliations
- PMID: 32571838
- PMCID: PMC7319273
- DOI: 10.1126/science.abc6952
A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2
Xiangyang Chi et al. Science. 2020.
Abstract
Developing therapeutics against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could be guided by the distribution of epitopes, not only on the receptor binding domain (RBD) of the Spike (S) protein but also across the full Spike (S) protein. We isolated and characterized monoclonal antibodies (mAbs) from 10 convalescent COVID-19 patients. Three mAbs showed neutralizing activities against authentic SARS-CoV-2. One mAb, named 4A8, exhibits high neutralization potency against both authentic and pseudotyped SARS-CoV-2 but does not bind the RBD. We defined the epitope of 4A8 as the N-terminal domain (NTD) of the S protein by determining with cryo-eletron microscopy its structure in complex with the S protein to an overall resolution of 3.1 angstroms and local resolution of 3.3 angstroms for the 4A8-NTD interface. This points to the NTD as a promising target for therapeutic mAbs against COVID-19.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
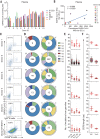
Fig. 1. Isolation of antigen-specific mAbs from convalescent patients of SARS-CoV-2.
(A) Reactions of plasma to SARS-CoV-2 proteins. S-ECD (extracellular domain of S protein), S1, S2, RBD (receptor binding domain), and N protein were used in ELISA to test the binding of plasma. Plasma of heathy donors were used as control, and cut-off values were calculated as optical density (OD) 450 of control × 2.1. Data were shown with mean and SD of a representative experiment. (B) The correlations between the authentic SARS-CoV-2 neutralizing antibody (NAb) titers and the pseudotyped SARS-CoV-2 NAb titers in plasma. Neutralizing assays of plasma against authentic SARS-CoV-2 were performed by using Vero E6 cells, and neutralization against pseudotyped SARS-CoV-2 were determined by using ACE2-293T cells. The correlations were calculated by means of Pearson correlation test in Graphpad 7.0. (C) Flow cytometry sorting from PBMCs of 10 convalescent patients. (D) Distribution of V gene families in heavy and light chains of all distinct clones (the total number is shown in the center of the pie charts) for each donor. (E) The number of amino acid (AA) and total nucleotide (Nt) mutations from the germline of all clonal sequences identified in (D) is shown. (F) CDR3 amino acid lengths of VH and VL of all clonal sequences identified in (D).
Fig. 2. Binding profiles of Spike protein–specific mAbs.
(A) Heatmap showing the binding of mAbs to different types of spike proteins determined by using ELISA. The EC50 value for each S-mAb combination is shown, with dark red, orange, yellow, or white shading indicating high, intermediate, low, or no detectable binding, respectively. EC50 values greater than 10,000 ng/ml are indicated (>). (B) Binding curves of representative mAbs. CR3022 is a control that was reported to bind SARS-CoV and SARS-CoV-2 RBD. Data were shown with mean and SD of a representative experiment. (C) Heatmap showing the competing binding of some representative S-reactive mAbs assayed in ELISA. Numbers in the box indicate the percentage binding of detecting mAb in the presence of the blocking antibody compared with the binding of detecting mAb in the absence of the blocking antibody. The mAbs were considered competing if the inhibiting percentage is <30% (black boxes with white numbers). The mAbs were judged to noncompete for the same site if the percentage is >70% (white boxes with red numbers). Gray boxes with black numbers indicate an intermediate phenotype (30 to ~70%). (D) Phylogenetic trees of all the S-specific mAbs.
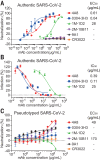
Fig. 3. Neutralizing capacities of S-reactive mAbs.
(A) Neutralization of S-reactive mAbs to authentic SARS-CoV-2 in Vero-E6 cells. (B) The authentic SARS-CoV-2 virus RNA load was determined in Vero-E6 cells treated with S-reactive mAbs by using quantitative PCR. Percent infection was calculated as the ratio of RNA load in mAb-treated wells to that in wells containing virus only. (C) Neutralization of S-reactive mAbs against HIV-vectored pseudotyped SARS-CoV-2 in ACE2-293T cells. Data were shown as mean ± SD of a representative experiment.
Fig. 4. 4A8 did not block the binding of Spike protein to ACE2 receptor.
(A) BLI sensorgrams and kinetics of mAbs binding to S proteins. Global fitting curves are shown as black lines. The _K_d were calculated by using a 1:1 binding model in Data Analysis Software 9.0, except for 2M-10B11, which used a heterogeneous ligand model owing to avidity effect. (B) The binding of S protein to human ACE2-overexpressing 293T cells were determined by means of flow cytometry. After the preincubation of S protein with each indicated mAb, the mAb-S mixtures were added to the ACE2-expressing cells. Cells were stained with anti-human IgG fluorescein isothiocyanate (mAb binding, x axis) and anti-His (S binding, y axis). Percentages of double-positive cells are shown. Control mAb CR3022 and 1A8 were previously reported to bind SARS-CoV RBD and Marburg glycoprotein, respectively, and ACE2-Fc protein was a human ACE2 protein conjugated with human Fc.
Fig. 5. Cryo-EM structure of the 4A8 and S-ECD complex.
The domain-colored cryo-EM map of the complex is shown on the left, and two perpendicular views of the overall structure are shown on the right. The heavy and light chains of 4A8 are colored blue and magenta, respectively. The NTDs of the trimeric S protein are colored orange. The one “up” RBD and two “down” RBDs of trimeric S protein are colored green and cyan, respectively.
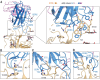
Fig. 6. Interactions between the NTD and 4A8.
(A) Extensive hydrophilic interactions on the interface between NTD and 4A8. Only one NTD-4A8 is shown. (B to D) Detailed analysis of the interface between NTD and 4A8. Polar interactions are indicated by red dashed lines. The residues involved in hydrophobic interactions are presented as spheres.
Similar articles
- Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. Pinto D, et al. Nature. 2020 Jul;583(7815):290-295. doi: 10.1038/s41586-020-2349-y. Epub 2020 May 18. Nature. 2020. PMID: 32422645 - Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JF, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen KY, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, Ho DD. Liu L, et al. Nature. 2020 Aug;584(7821):450-456. doi: 10.1038/s41586-020-2571-7. Epub 2020 Jul 22. Nature. 2020. PMID: 32698192 - Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.
Hussain A, Hasan A, Nejadi Babadaei MM, Bloukh SH, Chowdhury MEH, Sharifi M, Haghighat S, Falahati M. Hussain A, et al. Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review. - Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation.
Seydoux E, Homad LJ, MacCamy AJ, Parks KR, Hurlburt NK, Jennewein MF, Akins NR, Stuart AB, Wan YH, Feng J, Whaley RE, Singh S, Boeckh M, Cohen KW, McElrath MJ, Englund JA, Chu HY, Pancera M, McGuire AT, Stamatatos L. Seydoux E, et al. Immunity. 2020 Jul 14;53(1):98-105.e5. doi: 10.1016/j.immuni.2020.06.001. Epub 2020 Jun 8. Immunity. 2020. PMID: 32561270 Free PMC article. - Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2.
Yu F, Xiang R, Deng X, Wang L, Yu Z, Tian S, Liang R, Li Y, Ying T, Jiang S. Yu F, et al. Signal Transduct Target Ther. 2020 Sep 22;5(1):212. doi: 10.1038/s41392-020-00318-0. Signal Transduct Target Ther. 2020. PMID: 32963228 Free PMC article. Review.
Cited by
- The Genetic Variant of SARS-CoV-2: would It Matter for Controlling the Devastating Pandemic?
Guo S, Liu K, Zheng J. Guo S, et al. Int J Biol Sci. 2021 Apr 10;17(6):1476-1485. doi: 10.7150/ijbs.59137. eCollection 2021. Int J Biol Sci. 2021. PMID: 33907511 Free PMC article. Review. - Homologous and heterologous serological response to the N-terminal domain of SARS-CoV-2 in humans and mice.
Lv H, Tsang OT, So RTY, Wang Y, Yuan M, Liu H, Yip GK, Teo QW, Lin Y, Liang W, Wang J, Ng WW, Wilson IA, Peiris JSM, Wu NC, Mok CKP. Lv H, et al. Eur J Immunol. 2021 Sep;51(9):2296-2305. doi: 10.1002/eji.202149234. Epub 2021 Jun 22. Eur J Immunol. 2021. PMID: 34089541 Free PMC article. - SARS-CoV-2 protein subunit vaccination of mice and rhesus macaques elicits potent and durable neutralizing antibody responses.
Mandolesi M, Sheward DJ, Hanke L, Ma J, Pushparaj P, Perez Vidakovics L, Kim C, Àdori M, Lenart K, Loré K, Castro Dopico X, Coquet JM, McInerney GM, Karlsson Hedestam GB, Murrell B. Mandolesi M, et al. Cell Rep Med. 2021 Apr 20;2(4):100252. doi: 10.1016/j.xcrm.2021.100252. Epub 2021 Apr 5. Cell Rep Med. 2021. PMID: 33842900 Free PMC article. - Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial.
Zhu FC, Guan XH, Li YH, Huang JY, Jiang T, Hou LH, Li JX, Yang BF, Wang L, Wang WJ, Wu SP, Wang Z, Wu XH, Xu JJ, Zhang Z, Jia SY, Wang BS, Hu Y, Liu JJ, Zhang J, Qian XA, Li Q, Pan HX, Jiang HD, Deng P, Gou JB, Wang XW, Wang XH, Chen W. Zhu FC, et al. Lancet. 2020 Aug 15;396(10249):479-488. doi: 10.1016/S0140-6736(20)31605-6. Epub 2020 Jul 20. Lancet. 2020. PMID: 32702299 Free PMC article. Clinical Trial. - Nanobody Repertoires for Exposing Vulnerabilities of SARS-CoV-2.
Mast FD, Fridy PC, Ketaren NE, Wang J, Jacobs EY, Olivier JP, Sanyal T, Molloy KR, Schmidt F, Rutkowska M, Weisblum Y, Rich LM, Vanderwall ER, Dambrauskas N, Vigdorovich V, Keegan S, Jiler JB, Stein ME, Olinares PDB, Hatziioannou T, Sather DN, Debley JS, Fenyö D, Sali A, Bieniasz PD, Aitchison JD, Chait BT, Rout MP. Mast FD, et al. bioRxiv [Preprint]. 2021 Apr 10:2021.04.08.438911. doi: 10.1101/2021.04.08.438911. bioRxiv. 2021. PMID: 33851164 Free PMC article. Updated. Preprint.
References
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). 10.1016/S0140-6736(20)30183-5 - DOI - PMC - PubMed
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous