The E2 Marie Kondo and the CTLH E3 ligase clear deposited RNA binding proteins during the maternal-to-zygotic transition - PubMed (original) (raw)
The E2 Marie Kondo and the CTLH E3 ligase clear deposited RNA binding proteins during the maternal-to-zygotic transition
Michael Zavortink et al. Elife. 2020.
Abstract
The maternal-to-zygotic transition (MZT) is a conserved step in animal development, where control is passed from the maternal to the zygotic genome. Although the MZT is typically considered from its impact on the transcriptome, we previously found that three maternally deposited Drosophila RNA-binding proteins (ME31B, Trailer Hitch [TRAL], and Cup) are also cleared during the MZT by unknown mechanisms. Here, we show that these proteins are degraded by the ubiquitin-proteasome system. Marie Kondo, an E2 conjugating enzyme, and the E3 CTLH ligase are required for the destruction of ME31B, TRAL, and Cup. Structure modeling of the Drosophila CTLH complex suggests that substrate recognition is different than orthologous complexes. Despite occurring hours earlier, egg activation mediates clearance of these proteins through the Pan Gu kinase, which stimulates translation of Kdo mRNA. Clearance of the maternal protein dowry thus appears to be a coordinated, but as-yet underappreciated, aspect of the MZT.
Keywords: D. melanogaster; RNA binding proteins; chromosomes; developmental biology; gene expression; maternal to zygotic transition; post-transcriptional regulation; ubiquitin-proteasome system.
Plain language summary
Bestselling author and organizing consultant Marie Kondo has helped people around the world declutter their homes by getting rid of physical items that do not bring them joy. Keeping the crowded environment inside a living cell organized also requires work and involves removing molecules that are no longer needed. A fertilized egg cell, for example, contains molecules from the mother that regulate the initial stages as it develops into an embryo. Later on, the embryo takes control of its own development by destroying these inherited molecules and switches to making its own instead. This process is called the maternal-to-zygotic transition. The molecules passed from the mother to the egg cell include proteins and messenger RNAs (molecules that include the coded instructions to make new proteins). Previous research has begun to reveal how the embryo destroys the mRNAs it inherits from its mother and how it starts to make its own. Yet almost nothing is known about how an embryo gets rid of its mother’s proteins. To address this question, Zavortink, Rutt, Dzitoyeva et al. used an approach known as an RNA interference screen to identify factors required to destroy three maternal proteins in fruit fly embryos. The experiments helped identify one enzyme that worked together with another larger enzyme complex to destroy the maternal proteins. This enzyme belongs to a class of enzymes known as ubiquitin-conjugating enzymes (or E2 enzymes) and it was given the name “Kdo”, short for “Marie Kondo”. Further experiments showed that the mRNAs that code for the Kdo enzyme were present in unfertilized eggs, but in a repressed state that prevented the eggs from making the enzyme. Once an egg started to develop into an embryo, these mRNAs became active and the embryo started to make Kdo enzymes. This led to the three maternal proteins being destroyed during the maternal-to-zygotic transition. These findings reveal a new pathway that regulates the destruction of maternal proteins as the embryo develops. The next challenge will be identifying other maternal proteins that do not “spark joy” and understanding the role their destruction plays in the earliest events of embryonic development.
© 2020, Zavortink et al.
Conflict of interest statement
MZ, LR, SD, JH, CB, DB, MW, XC, OR No competing interests declared
Figures
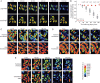
Figure 1.. The PNG kinase, but not fertilization, is required for ME31B degradation.
(A) PNG is required for the destruction of ME31B. Embryos from female flies of the indicated genotypes (that also contained the ME31B-GFP allele) were visualized at various time points after egg laying. Fluorescent images are false-colored so that more intense fluorescence is indicated by hotter colors; fluorescence scale is shown at bottom of figure. (B) ME31B-GFP is stabilized in png mutants. ME31B-GFP fluorescence from (A) was quantified. For each embryo, the fluorescence was normalized to its intensity at 0 min, and the fraction remaining is plotted through the time course. Significance was calculated using the Mann-Whitney test. (C) GNU is required for the destruction of ME31B. dsRNA targeting GNU or mCherry (as a control) mRNA was expressed during oogenesis; female flies also contained the ME31B-GFP allele. Laid embryos were visualized by fluorescence microscopy at indicated time points, otherwise as in A. (D) PLU is required for the destruction of ME31B. As in C, except with dsRNA targeting PLU mRNA. (E) Fertilization is not required for the destruction of ME31B. As in A, except with fertilized and unfertilized embryos.
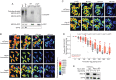
Figure 2.. ME31B is degraded by the ubiquitin-proteasome pathway.
(A) Ubiquitination of ME31B-GFP requires the PNG kinase. ME31B-GFP was immunoprecipitated under stringent conditions from 1–2 hr lysates from wild-type or png50 embryos, and then analyzed by western blotting by probing with α-GFP, α-eIF4E or α-ubiquitin. (B) ME31B-GFP is not degraded by autophagy. Embryos from female flies expressing the indicated dsRNAs and ME31B-GFP were imaged at various time points by fluorescence microscopy. Fluorescent images are false-colored so that more intense fluorescence is indicated by hotter colors; fluorescence scale is shown. (C) Inhibiting the proteasome partially stabilizes ME31B-GFP. As in B, except for dsRNAs targeting mCherry or Rpn10, a proteasome component. (D) ME31B-GFP is stabilized in Rpn10-depleted embryos. ME31B-GFP fluorescence from (C) and intervening time points was quantified. For each embryo, the fluorescence was normalized to its intensity at 0 min, and the fraction remaining is plotted through the time course. Significance was calculated using the Mann-Whitney test. (E) Depleting the proteasome partially stabilizes ME31B-GFP. Staged embryos from the indicated times were harvested with mCherry or Rpn11 knocked down. Western blotting was performed on the lysates, probing for GFP and eIF4E (as a loading control). Related to Source Figure 2—source data 1.
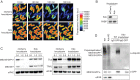
Figure 3.. The Marie Kondo E2 conjugating enzyme mediates the degradation of ME31B, TRAL, and Cup.
(A) Marie Kondo (Kdo/UBC-E2H) is required for the degradation of ME31B-GFP. Embryos laid from female flies expressing the indicated dsRNAs and ME31B-GFP were imaged at various time points by fluorescence microscopy. Fluorescent images are false-colored so that more intense fluorescence is indicated by hotter colors; fluorescence scale is shown. (B) Kdo is depleted in knockdown lines. An antibody was raised against Kdo, and 1–2 hr embryo lysates with the indicated dsRNA were probed for Kdo and PABP (as a loading control). (C) Kdo is required for destruction of ME31B, TRAL, and Cup. Staged embryos with mCherry or Kdo knocked down were harvested at the indicated times. Western blotting was performed on the lysates, probing for ME31B, Cup, TRAL, and eIF4E (as a loading control). Because the maternal flies contained wild-type and ME31B-GFP alleles, both proteins are visible in the α-ME31B blot. (D) Kdo is required for the ubiquitination of ME31B. ME31B-GFP was immunoprecipitated from 1 to 2 hr mCherry or Kdo knock-down embryo lysates under stringent conditions and then analyzed by western blotting by probing with α-GFP, α-eIF4E, or α-ubiquitin. IgG was used as an immunoprecipitation control.
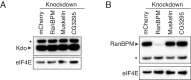
Figure 4.. The CTLH E3 ligase mediates the degradation of ME31B, TRAL, and Cup.
(A) Putative components of the Drosophila CTLH complex. Drosophila orthologs of the human components of the CTLH complex were determined by BLAST. Some components, such as GID4, appear to not have an identifiable ortholog. (B) ME31B interacts with the CTLH complex. Proteins interacting with ME31B-GFP at 1–2 and 2–3 hr after egg laying were determined by immunoprecipitation with α-GFP and mass spectrometry. Plotted are number of peptides detected (after subtracting for the signal in the control IgG immunoprecipitation). Known interactors with ME31B, such as Cup and TRAL, are shown in blue; CTLH components and ubiquitin are shown in orange; other hits from the screen are shown in brown. (C) Cup interacts with the CTLH complex. As in B, except for Cup-GFP. (D) The CTLH complex is required for destruction of ME31B-GFP. Embryos laid from the indicated maternal RNAi lines were visualized for 3 hr after egg-laying for ME31B-GFP fluorescence. Fluorescent images are false-colored so that more intense fluorescence is indicated by hotter colors; fluorescence scale is shown. (E) The CTLH complex is required for destruction of ME31B, TRAL, and Cup. Staged embryos from the indicated times were harvested with mCherry, RanBPM, Muskelin (Musk.), or CG3295 knocked down. Western blotting was performed on the lysates, probing for ME31B, Cup, TRAL, and eIF4E (as a loading control). Because the maternal flies contained wild-type and ME31B-GFP alleles, both proteins are visible in the α-ME31B blot. (F) ME31B-GFP interacts with RanBPM, a component of the CTLH complex. ME31B-GFP complexes were immunoprecipitated from 1 to 2 hr embryo lysates using α-GFP or IgG (as a control). Membranes were probed with α-GFP or α-RanBPM. *, non-specific band.
Figure 4—figure supplement 1.. Depletion of CTLH components does not impact Kdo levels.
(A) Kdo levels are not affected by CTLH complex depletion. Lysates from 0 to 2 hr embryos with the indicated knockdowns were analyzed by western blotting, probing for Kdo and eIF4E. * indicates a non-specific band. (B) Depletion of Muskelin and CG3295 does not affect RanBPM levels. As in A, except probing for RanBPM and eIF4E.
Figure 5.. The Drosophila CTLH complex has a different organization than the S. cerevisiae Gid complex.
(A) Schematic of the S. cerevisiae Gid complex based on recent cryo-EM structures (Qiao et al., 2020), and the Drosophila CTLH complex, based on orthology of the subunits. (B) The interaction between ME31B-GFP and RanBPM depends on Muskelin, but not Souji. ME31B-GFP complexes were immunoprecipitated from 1 to 2 hr embryo lysates using α-GFP or IgG (as a control). Immunoprecipitations were performed in 1–2 hr embryo lysates with mCherry, RanBPM, Muskelin, or Souji knocked-down. Membranes were probed with α-GFP or α-RanBPM. * indicates a non-specific band. (C) RanBPM and Hou share general structural similarities with their yeast orthologs. RanBPM (teal) and Hou (magenta) were threaded through structures of their orthologs, Gid1 (blue) and Gid8 (red), respectively. (D) RanBPM lacks the Gid4-interacting loop. Shown is the region of the S. cerevisiae Gid complex (left) and D. melanogaster CTLH complex (right) where Gid1 interacts with Gid4. The corresponding loop is missing in RanBPM. (E) Houki lacks the Gid5-interacting tail. Shown is the region of the S. cerevisiae Gid complex (left) and D. melanogaster CTLH complex (right) where Gid8 interacts with Gid5. The corresponding tail is missing in Houki. For simplicity, only Gid5 and Gid8 are shown.
Figure 5—figure supplement 1.. RanBPM and Houki lack key residues for CTLH complex formation.
(A) RanBPM and Gid1 protein sequence alignments. Shown is the sequence alignment for RanBPM with the residues around the Gid1 loop that interacts with Gid4. The known and predicted secondary structures are shown as determined by Phyre. b, β sheet; S, bend;. , missing residue. (B) Houki and Gid8 protein sequence alignments. Shown is the sequence alignment for Houki with the residues in the Gid8 tail that interacts with Gid5. b, β sheet; S, bend; a, α helix; T, hydrogen-bonded turn;. , missing residue.
Figure 6.. The interaction between ME31B and the CTLH complex does not depend on PNG.
(A) The interaction between ME31B-GFP and RanBPM does not depend on Kdo. ME31B-GFP complexes were immunoprecipitated from 1–2 hr embryo lysates using α-GFP or IgG (as a control). Immunoprecipitations were performed in 1–2 hr embryo lysates with Kdo knocked down. *, non-specific band. (B) The interaction between ME31B-GFP and RanBPM does not depend on PNG. As in B, except in 1–2 hr png50 embryo lysates. *, non-specific band. (C) The interaction between Cup and RanBPM does not depend on PNG. Complexes were immunoprecipitated from 1–2 hr lysates from wild-type (w1118) or png50 embryos using α-Cup or IgG. Membranes were probed with α-Cup or α-RanBPM. *, non-specific band.
Figure 7.. The PNG kinase mediates translational upregulation of Kdo at egg activation.
(A) Translation of Kdo increases at egg activation. Translational efficiency of Kdo mRNA was measured in published ribosome profiling datasets (Eichhorn et al., 2016). Shown is the translational efficiency in stage 11, 12, 13, and 14 oocytes, and 0–1 hr embryos. (B) Translational upregulation of Kdo depends on PNG. As in A, except for png50 stage 14 oocytes and activated eggs. (C) Translation of Kdo is highly dependent on PNG. Shown is a density plot of the difference in translational efficiency between wild-type and png50 embryos. The difference in translational efficiency for Kdo mRNA is shown by an arrow. (D) Production of Kdo during embryogenesis depends on PNG. Staged wild-type or png50 embryos were harvested at the indicated times after egg laying. Western blotting was performed on the lysates, probing for Kdo and PABP (as a loading control). (E) Kdo mRNA poly(A)-tail length increases at egg activation. As in A, except plotting poly(A)-tail lengths from Eichhorn, et al. (F) Lengthening of Kdo mRNA poly(A)-tail length depends on PNG. As in B, except plotting poly(A)-tail length. (G) Changes in poly(A)-tail length during egg activation correlate with changes in translation efficiency. Shown is scatter plot comparing, for each gene, the change in poly(A)-tail length between stage 14 oocytes and 0–1 hr embryos and the change in its translational efficiency. Kdo is highlighted in red. (H) ME31B changes upon egg activation. Shown is a scatter plot comparing ME31B binding to mRNAs in stage 14 oocytes and 0–1 hr embryos. Kdo is highlighted in red. (I) Changes in ME31B binding upon egg activation depend on PNG. As in H, except comparing ME31B binding in stage 14 oocytes and 0–1 hr png50 embryos. (J) Changes in ME31B at egg activation correlate with changes in translational efficiency. Shown is a scatter plot comparing changes in ME31B binding between stage 14 oocytes and 0–1 hr embryos and changes in translational efficiency. Kdo is highlighted in red.
Figure 7—figure supplement 1.. Translation of CTLH components does not increase during the oocyte-to-embryo transition.
(A) Translation of CTLH components does not increase at egg activation. Translational efficiency of CTLH components was measured in published ribosome profiling datasets (Eichhorn et al., 2016). Shown is the translational efficiency in stage 14 oocytes and 0–1 hr embryos. (B) Translation of CTLH components is not stimulated by the PNG kinase. As in B, except for png50 stage 14 oocytes and activated eggs.
Similar articles
- Precise Temporal Regulation of Post-transcriptional Repressors Is Required for an Orderly Drosophila Maternal-to-Zygotic Transition.
Cao WX, Kabelitz S, Gupta M, Yeung E, Lin S, Rammelt C, Ihling C, Pekovic F, Low TCH, Siddiqui NU, Cheng MHK, Angers S, Smibert CA, Wühr M, Wahle E, Lipshitz HD. Cao WX, et al. Cell Rep. 2020 Jun 23;31(12):107783. doi: 10.1016/j.celrep.2020.107783. Cell Rep. 2020. PMID: 32579915 Free PMC article. - Muskelin is a substrate adaptor of the highly regulated Drosophila embryonic CTLH E3 ligase.
Briney CA, Henriksen JC, Lin C, Jones LA, Benner L, Rains AB, Gutierrez R, Gafken PR, Rissland OS. Briney CA, et al. EMBO Rep. 2025 Mar;26(6):1647-1669. doi: 10.1038/s44319-025-00397-6. Epub 2025 Feb 20. EMBO Rep. 2025. PMID: 39979464 Free PMC article. - ME31B globally represses maternal mRNAs by two distinct mechanisms during the Drosophila maternal-to-zygotic transition.
Wang M, Ly M, Lugowski A, Laver JD, Lipshitz HD, Smibert CA, Rissland OS. Wang M, et al. Elife. 2017 Sep 6;6:e27891. doi: 10.7554/eLife.27891. Elife. 2017. PMID: 28875934 Free PMC article. - Regulatory principles governing the maternal-to-zygotic transition: insights from Drosophila melanogaster.
Hamm DC, Harrison MM. Hamm DC, et al. Open Biol. 2018 Dec;8(12):180183. doi: 10.1098/rsob.180183. Open Biol. 2018. PMID: 30977698 Free PMC article. Review. - Setting the stage for development: the maternal-to-zygotic transition in Drosophila.
Harrison MM, Marsh AJ, Rushlow CA. Harrison MM, et al. Genetics. 2023 Oct 4;225(2):iyad142. doi: 10.1093/genetics/iyad142. Genetics. 2023. PMID: 37616526 Free PMC article. Review.
Cited by
- Muskelin acts as a substrate receptor of the highly regulated Drosophila CTLH E3 ligase during the maternal-to-zygotic transition.
Briney CA, Henriksen JC, Lin C, Jones LA, Benner L, Rains AB, Gutierrez R, Gafken PR, Rissland OS. Briney CA, et al. bioRxiv [Preprint]. 2024 Jul 1:2024.06.28.601265. doi: 10.1101/2024.06.28.601265. bioRxiv. 2024. PMID: 39005399 Free PMC article. Preprint. - Skraban-Deardorff intellectual disability syndrome-associated mutations in WDR26 impair CTLH E3 complex assembly.
Gross A, Müller J, Chrustowicz J, Strasser A, Gottemukkala KV, Sherpa D, Schulman BA, Murray PJ, Alpi AF. Gross A, et al. FEBS Lett. 2024 May;598(9):978-994. doi: 10.1002/1873-3468.14866. Epub 2024 Apr 4. FEBS Lett. 2024. PMID: 38575527 Free PMC article. - How the ends signal the end: Regulation by E3 ubiquitin ligases recognizing protein termini.
Sherpa D, Chrustowicz J, Schulman BA. Sherpa D, et al. Mol Cell. 2022 Apr 21;82(8):1424-1438. doi: 10.1016/j.molcel.2022.02.004. Epub 2022 Mar 4. Mol Cell. 2022. PMID: 35247307 Free PMC article. Review. - Modular UBE2H-CTLH E2-E3 complexes regulate erythroid maturation.
Sherpa D, Mueller J, Karayel Ö, Xu P, Yao Y, Chrustowicz J, Gottemukkala KV, Baumann C, Gross A, Czarnecki O, Zhang W, Gu J, Nilvebrant J, Sidhu SS, Murray PJ, Mann M, Weiss MJ, Schulman BA, Alpi AF. Sherpa D, et al. Elife. 2022 Dec 2;11:e77937. doi: 10.7554/eLife.77937. Elife. 2022. PMID: 36459484 Free PMC article. - Specialized germline P-bodies are required to specify germ cell fate in Caenorhabditis elegans embryos.
Cassani M, Seydoux G. Cassani M, et al. Development. 2022 Nov 1;149(21):dev200920. doi: 10.1242/dev.200920. Epub 2022 Nov 7. Development. 2022. PMID: 36196602 Free PMC article.
References
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. - DOI - PMC - PubMed
- Cao WX, Kabelitz S, Gupta M, Yeung E, Lin S, Rammelt C, Ihling C, Pekovic F, Low TCH, Siddiqui NU, Cheung MHK, Angers S, Smibert CA, Wühr M, Wahle E, Lipshitz HD. Precise temporal regulation of post-transcriptional repressors is required for an orderly Drosophila maternal-to-zygotic transition. bioRxiv. 2019 doi: 10.1101/862490. - DOI - PMC - PubMed
- Chen L, Dumelie JG, Li X, Cheng MH, Yang Z, Laver JD, Siddiqui NU, Westwood JT, Morris Q, Lipshitz HD, Smibert CA. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biology. 2014;15:4–21. doi: 10.1186/gb-2014-15-1-r4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases