Targeting the ERG oncogene with splice-switching oligonucleotides as a novel therapeutic strategy in prostate cancer - PubMed (original) (raw)
. 2020 Sep;123(6):1024-1032.
doi: 10.1038/s41416-020-0951-2. Epub 2020 Jun 25.
Lisa Hobson 2, Laura Perry 2, Bethany Clark 2, Susan Heavey 3, Aiman Haider 4, Ashwin Sridhar 5, Greg Shaw 5, John Kelly 5, Alex Freeman 4, Ian Wilson 2, Hayley Whitaker 3, Elmar Nurmemmedov 6, Sebastian Oltean 1, Sean Porazinski # 7 8, Michael Ladomery # 9
Affiliations
- PMID: 32581342
- PMCID: PMC7493922
- DOI: 10.1038/s41416-020-0951-2
Targeting the ERG oncogene with splice-switching oligonucleotides as a novel therapeutic strategy in prostate cancer
Ling Li et al. Br J Cancer. 2020 Sep.
Abstract
Background: The ERG oncogene, a member of the ETS family of transcription factor encoding genes, is a genetic driver of prostate cancer. It is activated through a fusion with the androgen-responsive TMPRSS2 promoter in 50% of cases. There is therefore significant interest in developing novel therapeutic agents that target ERG. We have taken an antisense approach and designed morpholino-based oligonucleotides that target ERG by inducing skipping of its constitutive exon 4.
Methods: We designed antisense morpholino oligonucleotides (splice-switching oligonucleotides, SSOs) that target both the 5' and 3' splice sites of ERG's exon 4. We tested their efficacy in terms of inducing exon 4 skipping in two ERG-positive cell lines, VCaP prostate cancer cells and MG63 osteosarcoma cells. We measured their effect on cell proliferation, migration and apoptosis. We also tested their effect on xenograft tumour growth in mice and on ERG protein expression in a human prostate cancer radical prostatectomy sample ex vivo.
Results: In VCaP cells, both SSOs were effective at inducing exon 4 skipping, which resulted in a reduction of overall ERG protein levels up to 96 h following a single transfection. SSO-induced ERG reduction decreased cell proliferation, cell migration and significantly increased apoptosis. We observed a concomitant reduction in protein levels for cyclin D1, c-Myc and the Wnt signalling pathway member β-catenin as well as a marker of activated Wnt signalling, p-LRP6. We tested the 3' splice site SSO in MG63 xenografts in mice and observed a reduction in tumour growth. We also demonstrated that the 3' splice site SSO caused a reduction in ERG expression in a patient-derived prostate tumour tissue cultured ex vivo.
Conclusions: We have successfully designed and tested morpholino-based SSOs that cause a marked reduction in ERG expression, resulting in decreased cell proliferation, a reduced migratory phenotype and increased apoptosis. Our initial tests on mouse xenografts and a human prostate cancer radical prostatectomy specimen indicate that SSOs can be effective for oncogene targeting in vivo. As such, this study encourages further in vivo therapeutic studies using SSOs targeting the ERG oncogene.
Conflict of interest statement
The authors declare no competing interests.
Figures
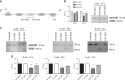
Fig. 1. Validation of splice-switching oligonucleotides targeting ERG exon 4 in VCaP cells.
a Schematic showing targeting of ERG exon 4 (E4) with splice-switching oligonucleotides (SSOs) designed against the 3′ (E43′) or 5′ (E45′) splice sites (see Supplementary Fig. 1a for additional SSO sequence details). Skipping of exon 4 is denoted by the dashed line. PCR primers to detect the presence or absence of exon 4 are indicated (2F and 6R). b Quantification of dose-dependent exon 4 skipping in VCaP cells treated with E4 SSOs for 24 h (n = 3, except for ctrl SSO at 6 µM, n = 2). A representative RT-PCR of exon 4 skipping is shown on the right. c Representative RT-PCR panels showing exon 4 skipping after 24–72 h treatment of VCaP cells with 8 µM E4 SSOs. d Quantifications of exon 4 skipping in VCaP cells treated with 8 µM E4 SSOs for 24–72 h (n = 3 at all timepoints). *** = p < 0.001, ** = p < 0.01, * = p < 0.05. Ctrl SSO control SSO.
Fig. 2. Splice-switching caused by ERG exon 4 SSOs reduces ERG protein levels in two cancer cell lines.
a Representative ERG and GAPDH (loading control) western blots of lysates from VCaP cells treated with 8 µM SSOs for 24, 72 and 96 h. The arrowhead indicates a putative truncated ERG isoform (full length is 54kD). b Quantifications of ERG western blots from VCaP cells treated with SSOs for 24–96 h (n = 4 for 24 h and 72 h, n = 3 for 96 h, except for ctrl SSO at 96 h, n = 2). c Representative RT-PCR panels for MG63 cells treated with 1 or 3 µM for 24 h. d Quantification of exon 4 skipping in MG63 cells treated with SSOs at 1 or 3 µM for 24 h (n = 3). e Representative ERG and β-actin (loading control) western blots of lysates from MG63 cells treated with SSOs for 48 and 72 h. The arrow indicates full-length ERG isoform (54 kD) and arrowhead indicates truncated ERG isoform. f Quantifications of ERG western blots from MG63 cells treated with 5 µM SSOs for 48 and 72 h (n = 8 for untreated and ctrl SSO, for E43′ n = 4 at 48 h; n = 6 at 72 h). ERG protein expression levels were normalised to GAPDH. *** = p < 0.001, ** = p < 0.01, * = p < 0.05. Ctrl SSO control SSO.
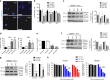
Fig. 3. ERG exon 4 SSOs affect VCaP cancer cell behaviour and signalling.
a Representative immunofluorescence images for Ki67 (grey) and Hoechst (blue) after 48 h of 8 µM E4 SSO treatment in VCaP cells. b Quantification of Ki67+ VCaP cells after 48–96 h of 8 µM E4 SSO treatment (n = 3 for all time points). c Western blotting and quantification for regulators of cell cycle progression, cyclin D1 and c-Myc, following 72 h of 8 µM E4 SSO treatment in VCaP cells (n = 3). β-actin was used as loading control. d Caspase-3/7 staining of VCaP cells treated with E4 SSOs at 8 µM for 48 h and 96 h (n = 3 at all timepoints). e Quantification of VCaP cells migrated in transwell assays after 48 h of E4 SSO treatment at 8 µM (n = 4, except for untreated where n = 3). f and g Representative western blotting for and quantification of key components of the canonical Wnt signalling pathway (β-catenin and p-LRP6) following 72 h of SSO treatment in VCaP cells (8 µM) and MG63 (3 µM) cells respectively (n = 3). β-actin was used as loading control. h TopFlash assays to assess Wnt pathway activity following 72 h of SSO treatment in VCaP cells and MG63 cells. *** = p < 0.001, ** = p < 0.01, * = p < 0.05. Unt untreated; Ctrl SSO control SSO. Scale bar = 40 µm.
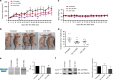
Fig. 4. ERG exon 4 SSOs have anti-tumour effects in vivo.
a Subcutaneous tumour growth of the ERG + MG63 cell line was measured twice weekly following IP injection of E43′ SSOs. Tumour measurements and dosing were performed on the same day. b Representative images of subcutaneous tumours (indicated by highlighted regions) in situ. c Measurements of body weight following systemic administration of SSOs. d Endpoint tumour weights from mice treated with SSOs (n = 8 for untreated; n = 11 for scrambled ctrl SSO; n = 10 for E43′ SSO). e Representative RT-PCR panels (left) and quantifications (right) of exon 4 skipping (arrow) from tumour tissue extracted at endpoints (n = 5 for untreated; n = 7 for scrambled ctrl and E43′ SSOs). f Representative panels for ERG and GAPDH (loading control) western blotting (left) in tumour tissue extracted from mice at endpoints. Quantification of ERG western blotting in tumours shown on right (n = 3 for untreated; n = 7 for scrambled ctrl SSO; n = 8 for E43′ SSO). ERG protein expression levels were normalised to GAPDH. *** = p < 0.001, ** = p < 0.01, * = p < 0.05. Scr ctrl SSO Scrambled control SSO.
Similar articles
- The Evolutionarily Conserved Cassette Exon 7b Drives ERG's Oncogenic Properties.
Jumbe SL, Porazinski SR, Oltean S, Mansell JP, Vahabi B, Wilson ID, Ladomery MR. Jumbe SL, et al. Transl Oncol. 2019 Jan;12(1):134-142. doi: 10.1016/j.tranon.2018.09.001. Epub 2018 Oct 5. Transl Oncol. 2019. PMID: 30296658 Free PMC article. - Identification of a Small Molecule That Selectively Inhibits ERG-Positive Cancer Cell Growth.
Mohamed AA, Xavier CP, Sukumar G, Tan SH, Ravindranath L, Seraj N, Kumar V, Sreenath T, McLeod DG, Petrovics G, Rosner IL, Srivastava M, Strovel J, Malhotra SV, LaRonde NA, Dobi A, Dalgard CL, Srivastava S. Mohamed AA, et al. Cancer Res. 2018 Jul 1;78(13):3659-3671. doi: 10.1158/0008-5472.CAN-17-2949. Epub 2018 Apr 30. Cancer Res. 2018. PMID: 29712692 - Role of dutasteride in pre-clinical ETS fusion-positive prostate cancer models.
Ateeq B, Vellaichamy A, Tomlins SA, Wang R, Cao Q, Lonigro RJ, Pienta KJ, Varambally S. Ateeq B, et al. Prostate. 2012 Oct 1;72(14):1542-9. doi: 10.1002/pros.22509. Epub 2012 Mar 13. Prostate. 2012. PMID: 22415461 - The Expression of Proto-Oncogene ETS-Related Gene (ERG) Plays a Central Role in the Oncogenic Mechanism Involved in the Development and Progression of Prostate Cancer.
Khosh Kish E, Choudhry M, Gamallat Y, Buharideen SM, D D, Bismar TA. Khosh Kish E, et al. Int J Mol Sci. 2022 Apr 26;23(9):4772. doi: 10.3390/ijms23094772. Int J Mol Sci. 2022. PMID: 35563163 Free PMC article. Review.
Cited by
- Steering research on mRNA splicing in cancer towards clinical translation.
Anczukow O, Allain FH, Angarola BL, Black DL, Brooks AN, Cheng C, Conesa A, Crosse EI, Eyras E, Guccione E, Lu SX, Neugebauer KM, Sehgal P, Song X, Tothova Z, Valcárcel J, Weeks KM, Yeo GW, Thomas-Tikhonenko A. Anczukow O, et al. Nat Rev Cancer. 2024 Dec;24(12):887-905. doi: 10.1038/s41568-024-00750-2. Epub 2024 Oct 9. Nat Rev Cancer. 2024. PMID: 39384951 Free PMC article. Review. - Poly(ADP-ribose) Polyremase-1 (PARP-1) Inhibition: A Promising Therapeutic Strategy for ETS-Expressing Tumours.
Legrand AJ, Choul-Li S, Villeret V, Aumercier M. Legrand AJ, et al. Int J Mol Sci. 2023 Aug 30;24(17):13454. doi: 10.3390/ijms241713454. Int J Mol Sci. 2023. PMID: 37686260 Free PMC article. Review. - How Driver Oncogenes Shape and Are Shaped by Alternative Splicing Mechanisms in Tumors.
Wojtyś W, Oroń M. Wojtyś W, et al. Cancers (Basel). 2023 May 26;15(11):2918. doi: 10.3390/cancers15112918. Cancers (Basel). 2023. PMID: 37296881 Free PMC article. Review. - Splice-switch oligonucleotide-based combinatorial platform prioritizes synthetic lethal targets CHK1 and BRD4 against MYC-driven hepatocellular carcinoma.
Thng DKH, Toh TB, Pigini P, Hooi L, Dan YY, Chow PK, Bonney GK, Rashid MBMA, Guccione E, Wee DKB, Chow EK. Thng DKH, et al. Bioeng Transl Med. 2022 Sep 3;8(1):e10363. doi: 10.1002/btm2.10363. eCollection 2023 Jan. Bioeng Transl Med. 2022. PMID: 36684069 Free PMC article. - The landscape and biological relevance of aberrant alternative splicing events in esophageal squamous cell carcinoma.
Wu Q, Zhang Y, An H, Sun W, Wang R, Liu M, Zhang K. Wu Q, et al. Oncogene. 2021 Jun;40(24):4184-4197. doi: 10.1038/s41388-021-01849-8. Epub 2021 Jun 2. Oncogene. 2021. PMID: 34079089
References
- Werner MH, et al. The solution structure of the human ETS1-DNA complex reveals a novel mode of binding and true side chain intercalation. Cell. 1995;83:761–771. - PubMed
- Rao VN, Papas TS, Reddy E. ERG, a human ets-related gene on chromosome 21: alternative splicing, polyadenylation, and translation. Science. 1987;237:635–639. - PubMed
- Adamo P, Ladomery MR. The oncogene ERG: a key factor in prostate cancer. Oncogene. 2015;35:403–414. - PubMed
- Rosen P, Sesterhenn IA, Brassell SA, McLeod DG, Srivastava S, Dobi A. Clinical potential of the ERG oncoprotein in prostate cancer. Nat. Rev. Urol. 2012;10:483–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials