Encapsulins-Bacterial Protein Nanocompartments: Structure, Properties, and Application - PubMed (original) (raw)
Review
Encapsulins-Bacterial Protein Nanocompartments: Structure, Properties, and Application
Anna N Gabashvili et al. Biomolecules. 2020.
Abstract
Recently, a new class of prokaryotic compartments, collectively called encapsulins or protein nanocompartments, has been discovered. The shell proteins of these structures self-organize to form icosahedral compartments with a diameter of 25-42 nm, while one or more cargo proteins with various functions can be encapsulated in the nanocompartment. Non-native cargo proteins can be loaded into nanocompartments and the surface of the shells can be further functionalized, which allows for developing targeted drug delivery systems or using encapsulins as contrast agents for magnetic resonance imaging. Since the genes encoding encapsulins can be integrated into the cell genome, encapsulins are attractive for investigation in various scientific fields, including biomedicine and nanotechnology.
Keywords: MRI; cell labeling; encapsulin; nanocompartment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Figure 1
Structural comparison of the capsomers/monomers and assembled capsid/encapsulins: (A) HK97 phage, (B) Thermotoga maritima encapsulin, (C) Myxococcus xanthus encapsulin, (D) Pyrococcus furiosus encapsulin. (PDB-ID: 2FT1, 3DKT, 4PT2, and 2E0Z, respectively).
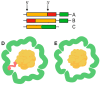
Figure 2
The cargo loading peptide coding sequence can be located at either the 5′ or 3′ end of the cargo loading peptide gene (A,B) with formation of the encapsulin structure (D). In some cases, the cargo protein encoding gene and encapsulin shell encoding gene are fused (C), and there is no need for a cargo-loading peptide to form encapsulin structure (E).
Similar articles
- Encapsulins: Structure, Properties, and Biotechnological Applications.
Chmelyuk NS, Oda VV, Gabashvili AN, Abakumov MA. Chmelyuk NS, et al. Biochemistry (Mosc). 2023 Jan;88(1):35-49. doi: 10.1134/S0006297923010042. Biochemistry (Mosc). 2023. PMID: 37068871 Free PMC article. Review. - Encapsulins: molecular biology of the shell.
Nichols RJ, Cassidy-Amstutz C, Chaijarasphong T, Savage DF. Nichols RJ, et al. Crit Rev Biochem Mol Biol. 2017 Oct;52(5):583-594. doi: 10.1080/10409238.2017.1337709. Epub 2017 Jun 21. Crit Rev Biochem Mol Biol. 2017. PMID: 28635326 Review. - Assembly and Mechanical Properties of the Cargo-Free and Cargo-Loaded Bacterial Nanocompartment Encapsulin.
Snijder J, Kononova O, Barbu IM, Uetrecht C, Rurup WF, Burnley RJ, Koay MS, Cornelissen JJ, Roos WH, Barsegov V, Wuite GJ, Heck AJ. Snijder J, et al. Biomacromolecules. 2016 Aug 8;17(8):2522-9. doi: 10.1021/acs.biomac.6b00469. Epub 2016 Jul 7. Biomacromolecules. 2016. PMID: 27355101 - Structure and heterogeneity of a highly cargo-loaded encapsulin shell.
Kwon S, Andreas MP, Giessen TW. Kwon S, et al. J Struct Biol. 2023 Dec;215(4):108022. doi: 10.1016/j.jsb.2023.108022. Epub 2023 Aug 30. J Struct Biol. 2023. PMID: 37657675 Free PMC article. - Encapsulation of Transketolase into _In Vitro_-Assembled Protein Nanocompartments Improves Thermal Stability.
Van de Steen A, Wilkinson HC, Dalby PA, Frank S. Van de Steen A, et al. ACS Appl Bio Mater. 2024 Jun 17;7(6):3660-3674. doi: 10.1021/acsabm.3c01153. Epub 2024 Jun 4. ACS Appl Bio Mater. 2024. PMID: 38835217 Free PMC article.
Cited by
- Protein nanocages: A new frontier in mucosal vaccine delivery and immune activation.
J LAA, Pa P, Seng CY, Rhee JH, Lee SE. J LAA, et al. Hum Vaccin Immunother. 2025 Dec;21(1):2492906. doi: 10.1080/21645515.2025.2492906. Epub 2025 May 12. Hum Vaccin Immunother. 2025. PMID: 40353600 Free PMC article. Review. - Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Tang Y, Mu A, Zhang Y, Zhou S, Wang W, Lai Y, Zhou X, Liu F, Yang X, Gong H, Wang Q, Rao Z. Tang Y, et al. Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2025658118. doi: 10.1073/pnas.2025658118. Proc Natl Acad Sci U S A. 2021. PMID: 33853951 Free PMC article. - Embedding a membrane protein into an enveloped artificial viral replica.
Furukawa H, Inaba H, Sasaki Y, Akiyoshi K, Matsuura K. Furukawa H, et al. RSC Chem Biol. 2021 Dec 21;3(2):231-241. doi: 10.1039/d1cb00166c. eCollection 2022 Feb 9. RSC Chem Biol. 2021. PMID: 35360888 Free PMC article. - Nanotechnological Applications Based on Bacterial Encapsulins.
Rodríguez JM, Allende-Ballestero C, Cornelissen JJLM, Castón JR. Rodríguez JM, et al. Nanomaterials (Basel). 2021 Jun 1;11(6):1467. doi: 10.3390/nano11061467. Nanomaterials (Basel). 2021. PMID: 34206092 Free PMC article. Review. - Myxococcus xanthus encapsulin cargo protein EncD is a flavin-binding protein with ferric reductase activity.
Eren E, Watts NR, Conway JF, Wingfield PT. Eren E, et al. Proc Natl Acad Sci U S A. 2024 May 21;121(21):e2400426121. doi: 10.1073/pnas.2400426121. Epub 2024 May 15. Proc Natl Acad Sci U S A. 2024. PMID: 38748579 Free PMC article.
References
- Rosenkrands I., Rasmussen P.B., Carnio M., Jacobsen S., Theisen M., Andersen P. Identification and Characterization of a 29-Kilodalton Protein from Mycobacterium Tuberculosis Culture Filtrate Recognized by Mouse Memory Effector Cells. Infect. Immun. 1998;66:2728–2735. doi: 10.1128/IAI.66.6.2728-2735.1998. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 19-45-06302/Russian Science Foundation/International
- K2-2019-011/Ministry of Education and Science of the Russian Federation/International
- Humboldt Research Fellowship for Postdoctoral Researchers/Alexander von Humboldt-Stiftung/International
LinkOut - more resources
Full Text Sources