NCBI Taxonomy: a comprehensive update on curation, resources and tools - PubMed (original) (raw)
Review
. 2020 Jan 1:2020:baaa062.
doi: 10.1093/database/baaa062.
Stacy Ciufo 1, Mikhail Domrachev 1, Carol L Hotton 1, Sivakumar Kannan 1, Rogneda Khovanskaya 1, Detlef Leipe 1, Richard Mcveigh 1, Kathleen O'Neill 1, Barbara Robbertse 1, Shobha Sharma 1, Vladimir Soussov 1, John P Sullivan 1, Lu Sun 1, Seán Turner 1, Ilene Karsch-Mizrachi 1
Affiliations
- PMID: 32761142
- PMCID: PMC7408187
- DOI: 10.1093/database/baaa062
Review
NCBI Taxonomy: a comprehensive update on curation, resources and tools
Conrad L Schoch et al. Database (Oxford). 2020.
Abstract
The National Center for Biotechnology Information (NCBI) Taxonomy includes organism names and classifications for every sequence in the nucleotide and protein sequence databases of the International Nucleotide Sequence Database Collaboration. Since the last review of this resource in 2012, it has undergone several improvements. Most notable is the shift from a single SQL database to a series of linked databases tied to a framework of data called NameBank. This means that relations among data elements can be adjusted in more detail, resulting in expanded annotation of synonyms, the ability to flag names with specific nomenclatural properties, enhanced tracking of publications tied to names and improved annotation of scientific authorities and types. Additionally, practices utilized by NCBI Taxonomy curators specific to major taxonomic groups are described, terms peculiar to NCBI Taxonomy are explained, external resources are acknowledged and updates to tools and other resources are documented. Database URL: https://www.ncbi.nlm.nih.gov/taxonomy.
Published by Oxford University Press 2020.
Figures
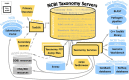
Figure 1
Summarized flow of NCBI Taxonomy information.
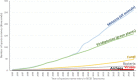
Figure 2
Species names added over time to NCBI Taxonomy. The first occurrence of each species in the NCBI Taxonomy was determined by the created date of its associated TaxNode. This date represents the first addition of the species into the database irrespective of subsequent name changes.
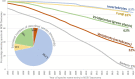
Figure 3
Estimate of the percentage of formal species names missing from the public NCBI databases. Curves were generated by plotting the number of formal species in the NCBI Taxonomy against the running total of described species in the corresponding group by the end of the year. The IJSEM was used as the source for bacteria. The International Plant Names Index (IPNI; 27) was used as the source for the green plants. The Species 2000 Annual Checklist (46) was used as the source for invertebrates and Fungi. Vertebrate data were collected from the Catalogue of Fishes (21), Amphibian Species of the World (17), the Reptile Database (32), Avibase (19) and the American Society of Mammalogists (18). Archaea and viruses were omitted for having a small number of species and a specialized process for reporting new species, respectively.
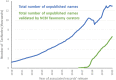
Figure 4
Total number of names labeled as unpublished in NCBI Taxonomy, over time.
Figure 5
NCBI TaxBrowser example page.
Similar articles
- Type material in the NCBI Taxonomy Database.
Federhen S. Federhen S. Nucleic Acids Res. 2015 Jan;43(Database issue):D1086-98. doi: 10.1093/nar/gku1127. Epub 2014 Nov 14. Nucleic Acids Res. 2015. PMID: 25398905 Free PMC article. - The NCBI Taxonomy database.
Federhen S. Federhen S. Nucleic Acids Res. 2012 Jan;40(Database issue):D136-43. doi: 10.1093/nar/gkr1178. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22139910 Free PMC article. - Database resources of the National Center for Biotechnology Information.
Sayers EW, Beck J, Bolton EE, Brister JR, Chan J, Comeau DC, Connor R, DiCuccio M, Farrell CM, Feldgarden M, Fine AM, Funk K, Hatcher E, Hoeppner M, Kane M, Kannan S, Katz KS, Kelly C, Klimke W, Kim S, Kimchi A, Landrum M, Lathrop S, Lu Z, Malheiro A, Marchler-Bauer A, Murphy TD, Phan L, Prasad AB, Pujar S, Sawyer A, Schmieder E, Schneider VA, Schoch CL, Sharma S, Thibaud-Nissen F, Trawick BW, Venkatapathi T, Wang J, Pruitt KD, Sherry ST. Sayers EW, et al. Nucleic Acids Res. 2024 Jan 5;52(D1):D33-D43. doi: 10.1093/nar/gkad1044. Nucleic Acids Res. 2024. PMID: 37994677 Free PMC article. - Database resources of the National Center for Biotechnology Information.
Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, Comeau DC, Funk K, Kim S, Klimke W, Marchler-Bauer A, Landrum M, Lathrop S, Lu Z, Madden TL, O'Leary N, Phan L, Rangwala SH, Schneider VA, Skripchenko Y, Wang J, Ye J, Trawick BW, Pruitt KD, Sherry ST. Sayers EW, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D10-D17. doi: 10.1093/nar/gkaa892. Nucleic Acids Res. 2021. PMID: 33095870 Free PMC article. Review. - Education resources of the National Center for Biotechnology Information.
Cooper PS, Lipshultz D, Matten WT, McGinnis SD, Pechous S, Romiti ML, Tao T, Valjavec-Gratian M, Sayers EW. Cooper PS, et al. Brief Bioinform. 2010 Nov;11(6):563-9. doi: 10.1093/bib/bbq022. Epub 2010 Jun 22. Brief Bioinform. 2010. PMID: 20570844 Free PMC article. Review.
Cited by
- Evolution of g-type lysozymes in metazoa: insights into immunity and digestive adaptations.
Mukherjee K, Moroz LL. Mukherjee K, et al. Front Cell Dev Biol. 2024 Nov 6;12:1487920. doi: 10.3389/fcell.2024.1487920. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39568508 Free PMC article. - The development of a lateral flow immunochromatographic test strip for measurement of specific IgA and IgG antibodies level against porcine epidemic diarrhea virus in pig milk.
Jermsutjarit P, Venkateswaran D, Indrawattana N, Na Plord J, Tantituvanont A, Nilubol D. Jermsutjarit P, et al. Vet Q. 2024 Dec;44(1):1-15. doi: 10.1080/01652176.2024.2429472. Epub 2024 Nov 21. Vet Q. 2024. PMID: 39568374 Free PMC article. - Emergency wound site infection caused by Gulosibacter massiliensis: a case report.
Li W, Zhang R, Liu L, Wang C, Sun Y, Dai Y, Yang X, Lin S. Li W, et al. BMC Infect Dis. 2024 Nov 13;24(1):1291. doi: 10.1186/s12879-024-10187-5. BMC Infect Dis. 2024. PMID: 39538156 Free PMC article. - A catalogue of chromosome counts for Phylum Nematoda.
Blaxter ML, Leech C, Lunt DH. Blaxter ML, et al. Wellcome Open Res. 2024 Feb 19;9:55. doi: 10.12688/wellcomeopenres.20550.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39534537 Free PMC article. - Taxonomy Identifiers (TaxId) for Biodiversity Genomics: a guide to getting TaxId for submission of data to public databases.
Blaxter M, Pauperio J, Schoch C, Howe K. Blaxter M, et al. Wellcome Open Res. 2024 Oct 15;9:591. doi: 10.12688/wellcomeopenres.22949.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39526195 Free PMC article.
References
- Strasser B.J. (2008) GenBank—natural history in the 21st century? Science, 322, 537–538. - PubMed
- Schuler G.D., Epstein J.A., Ohkawa H. et al. (1996) Entrez: molecular biology database and retrieval system. Methods Enzymol., 266, 141–162. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources