Genome-wide identification of genes regulating DNA methylation using genetic anchors for causal inference - PubMed (original) (raw)
doi: 10.1186/s13059-020-02114-z.
René Luijk 1, Lucia Daxinger 3, Maarten van Iterson 1, Koen F Dekkers 1, Rick Jansen 4; BIOS Consortium; Joyce B J van Meurs 5, Peter A C 't Hoen 6, M Arfan Ikram 7, Marleen M J van Greevenbroek 8 9, Dorret I Boomsma 10, P Eline Slagboom 1, Jan H Veldink 2, Erik W van Zwet 11, Bastiaan T Heijmans 12
Collaborators, Affiliations
- PMID: 32859263
- PMCID: PMC7453518
- DOI: 10.1186/s13059-020-02114-z
Genome-wide identification of genes regulating DNA methylation using genetic anchors for causal inference
Paul J Hop et al. Genome Biol. 2020.
Abstract
Background: DNA methylation is a key epigenetic modification in human development and disease, yet there is limited understanding of its highly coordinated regulation. Here, we identify 818 genes that affect DNA methylation patterns in blood using large-scale population genomics data.
Results: By employing genetic instruments as causal anchors, we establish directed associations between gene expression and distant DNA methylation levels, while ensuring specificity of the associations by correcting for linkage disequilibrium and pleiotropy among neighboring genes. The identified genes are enriched for transcription factors, of which many consistently increased or decreased DNA methylation levels at multiple CpG sites. In addition, we show that a substantial number of transcription factors affected DNA methylation at their experimentally determined binding sites. We also observe genes encoding proteins with heterogenous functions that have widespread effects on DNA methylation, e.g., NFKBIE, CDCA7(L), and NLRC5, and for several examples, we suggest plausible mechanisms underlying their effect on DNA methylation.
Conclusion: We report hundreds of genes that affect DNA methylation and provide key insights in the principles underlying epigenetic regulation.
Keywords: Causal inference; Chromatin; DNA methylation; Epigenetic regulation; Functional genomics; Genetic instrumental variable; Pleiotropy; Transcription factor.
Conflict of interest statement
The authors declare no competing interests.
Figures
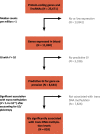
Fig. 1
Flowchart showing the successive steps leading to the identification of 818 genes that affect DNA methylation in trans
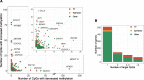
Fig. 2
A considerable fraction of the identified genes (N = 308) affected multiple target CpGs in trans. a Each dot represents a gene with trans DNA methylation effects. The _x_-axis shows the number of affected target CpGs with decreased methylation levels upon increased gene expression, and the _y_-axis shows the number of affected target CpGs with increased methylation levels upon increased gene expression. The figure in the right upper corner is a zoomed-in version in which only genes that affect less than 25 CpG sites in either direction are displayed. b Bars represent the number of genes with either 1, 2, 3–5, or more than 5 target CpGs. The percentage of genes that are annotated as transcription factors increases with the number of target CpGs
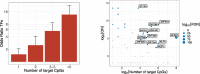
Fig. 3
a Enrichment (odds ratio) for transcription factors among identified genes with either 1, 2, 3–5, or more than 5 target CpGs. Error bars represent 95% confidence intervals. b Transcription factor binding site enrichment; each dot represents a transcription factor, with on the _x_-axis the logarithm of the number of target CpGs for that transcription factor (at a gene-level significance level; P < 1.2 × 10−7), and on the _y_-axis the odds ratio for the enrichment of the target CpGs in its experimentally determined binding sites (ChIP-seq). The size of the dots represents the significance (FDR), and TFs for which the target CpGs were significantly enriched in its binding sites are colored in blue
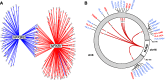
Fig. 4
a Network for transcription factor NFKB1 and its inhibitor NFKBIE. Gray circles indicate target CpGs, and arrows represent directed associations (i.e., association between GI and DNA methylation levels). Blue lines indicate a positive association between gene expression and DNA methylation levels; red lines indicate a negative association between gene expression levels and DNA methylation levels. b NLRC5 (chromosome 16) was associated with decreased DNA methylation levels at multiple (N = 43) CpG sites in the classical and extended MHC region (chromosome 6). Red lines indicate a negative association between NLRC5 expression levels and DNA methylation levels. The numbers displayed in the lines indicate how many target CpGs the line represents. Gene labels are displayed if one or more target CpGs were associated with the expression of these genes. Blue gene symbols refer to genes negatively correlated with target CpG methylation (implying upregulation by NLRC5), and vice versa for red labels. Asterisks indicate that the GI corresponding to NLRC5 was also (positively) associated with this gene
Fig. 5
a CDCA7 (located on chromosome 2) and CDCA7L (located on chromosome 7) both affect genome-wide DNA methylation levels. Blue lines indicate a positive association between CDCA7 expression and trans DNA methylation levels. Green lines indicate a positive association between CDCA7L expression levels and trans DNA methylation levels. b, c Over- or underrepresentation of target CpGs in predicted chromatin states for b CDCA7 and c CDCA7L. Blue bars represent enrichment of CpGs that are significant at a genome-wide significance level (P < 1.4 × 10−11), and gray bars represent enrichment of CpGs that are significant at a gene-level significance level (P < 1.2 × 10−7). BivFlnk, flanking bivalent TSS/enhancer; Enh, enhancer; EnhBiv, bivalent enhancer; EnhG, genic enhancer; Het, heterochromatin; Quies, quiescent; ReprPC, repressed polycomb; ReprPCWk, weak repressed polycomb; TssA, active TSS; TssAFlnk, flanking active TSS; TssBiv, bivalent/poised TSS; Tx, strong transcription; TxFlnk, weak transcription; ZNF/Rpts, ZNF genes and repeats
Similar articles
- Genome-Wide DNA Methylation and Gene Expression in Patients with Indolent Systemic Mastocytosis.
Górska A, Urbanowicz M, Grochowalski Ł, Seweryn M, Sobalska-Kwapis M, Wojdacz T, Lange M, Gruchała-Niedoszytko M, Jarczak J, Strapagiel D, Górska-Ponikowska M, Pelikant-Małecka I, Kalinowski L, Nedoszytko B, Gutowska-Owsiak D, Niedoszytko M. Górska A, et al. Int J Mol Sci. 2023 Sep 10;24(18):13910. doi: 10.3390/ijms241813910. Int J Mol Sci. 2023. PMID: 37762215 Free PMC article. - Methylation patterns associated with C-reactive protein in racially and ethnically diverse populations.
Lundin JI, Peters U, Hu Y, Ammous F, Avery CL, Benjamin EJ, Bis JC, Brody JA, Carlson C, Cushman M, Gignoux C, Guo X, Haessler J, Haiman C, Joehanes R, Kasela S, Kenny E, Lapalainien T, Levy D, Liu C, Liu Y, Loos RJF, Lu A, Matise T, North KE, Park SL, Ratliff SM, Reiner A, Rich SS, Rotter JI, Smith JA, Sotoodehnia N, Tracy R, Van den Berg D, Xu H, Ye T, Zhao W, Raffield LM, Kooperberg C; PAGE Study. Lundin JI, et al. Epigenetics. 2024 Dec;19(1):2333668. doi: 10.1080/15592294.2024.2333668. Epub 2024 Apr 3. Epigenetics. 2024. PMID: 38571307 Free PMC article. - Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets.
Olsson AH, Volkov P, Bacos K, Dayeh T, Hall E, Nilsson EA, Ladenvall C, Rönn T, Ling C. Olsson AH, et al. PLoS Genet. 2014 Nov 6;10(11):e1004735. doi: 10.1371/journal.pgen.1004735. eCollection 2014 Nov. PLoS Genet. 2014. PMID: 25375650 Free PMC article. - Epigenomics in stress tolerance of plants under the climate change.
Kumar M, Rani K. Kumar M, et al. Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review. - Epigenetic regulation in human melanoma: past and future.
Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Sarkar D, et al. Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review.
Cited by
- Pleiotropic influence of DNA methylation QTLs on physiological and ageing traits.
Mozhui K, Kim H, Villani F, Haghani A, Sen S, Horvath S. Mozhui K, et al. Epigenetics. 2023 Dec;18(1):2252631. doi: 10.1080/15592294.2023.2252631. Epigenetics. 2023. PMID: 37691384 Free PMC article. - Linking Prenatal Environmental Exposures to Lifetime Health with Epigenome-Wide Association Studies: State-of-the-Science Review and Future Recommendations.
Bakulski KM, Blostein F, London SJ. Bakulski KM, et al. Environ Health Perspect. 2023 Dec;131(12):126001. doi: 10.1289/EHP12956. Epub 2023 Dec 4. Environ Health Perspect. 2023. PMID: 38048101 Free PMC article. Review. - Role of Main RNA Methylation in Hepatocellular Carcinoma: N6-Methyladenosine, 5-Methylcytosine, and N1-Methyladenosine.
Xu Y, Zhang M, Zhang Q, Yu X, Sun Z, He Y, Guo W. Xu Y, et al. Front Cell Dev Biol. 2021 Nov 30;9:767668. doi: 10.3389/fcell.2021.767668. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34917614 Free PMC article. Review. - Developmental Dyslexia: Insights from EEG-Based Findings and Molecular Signatures-A Pilot Study.
Theodoridou D, Tsiantis CO, Vlaikou AM, Chondrou V, Zakopoulou V, Christodoulides P, Oikonomou ED, Tzimourta KD, Kostoulas C, Tzallas AT, Tsamis KI, Peschos D, Sgourou A, Filiou MD, Syrrou M. Theodoridou D, et al. Brain Sci. 2024 Jan 28;14(2):139. doi: 10.3390/brainsci14020139. Brain Sci. 2024. PMID: 38391714 Free PMC article. - Genetic variation influencing DNA methylation provides insights into molecular mechanisms regulating genomic function.
Hawe JS, Wilson R, Schmid KT, Zhou L, Lakshmanan LN, Lehne BC, Kühnel B, Scott WR, Wielscher M, Yew YW, Baumbach C, Lee DP, Marouli E, Bernard M, Pfeiffer L, Matías-García PR, Autio MI, Bourgeois S, Herder C, Karhunen V, Meitinger T, Prokisch H, Rathmann W, Roden M, Sebert S, Shin J, Strauch K, Zhang W, Tan WLW, Hauck SM, Merl-Pham J, Grallert H, Barbosa EGV; MuTHER Consortium; Illig T, Peters A, Paus T, Pausova Z, Deloukas P, Foo RSY, Jarvelin MR, Kooner JS, Loh M, Heinig M, Gieger C, Waldenberger M, Chambers JC. Hawe JS, et al. Nat Genet. 2022 Jan;54(1):18-29. doi: 10.1038/s41588-021-00969-x. Epub 2022 Jan 3. Nat Genet. 2022. PMID: 34980917 Free PMC article.
References
- Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777–786. - PubMed
- Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–266. - PubMed
- Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources