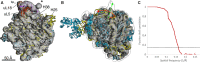Cryo-electron microscopy visualization of a large insertion in the 5S ribosomal RNA of the extremely halophilic archaeon Halococcus morrhuae - PubMed (original) (raw)
Cryo-electron microscopy visualization of a large insertion in the 5S ribosomal RNA of the extremely halophilic archaeon Halococcus morrhuae
Madhan R Tirumalai et al. FEBS Open Bio. 2020 Oct.
Abstract
The extreme halophile Halococcus morrhuae (ATCC® 17082) contains a 108-nucleotide insertion in its 5S rRNA. Large rRNA expansions in Archaea are rare. This one almost doubles the length of the 5S rRNA. In order to understand how such an insertion is accommodated in the ribosome, we obtained a cryo-electron microscopy reconstruction of the native large subunit at subnanometer resolution. The insertion site forms a four-way junction that fully preserves the canonical 5S rRNA structure. Moving away from the junction site, the inserted region is conformationally flexible and does not pack tightly against the large subunit. The high-salt requirement of the H. morrhuae ribosomes for their stability conflicted with the low-salt threshold for cryo-electron microscopy procedures. Despite this obstacle, this is the first cryo-electron microscopy map of Halococcus ribosomes.
Keywords: accretion model; archaea; expansion sequences; insertion sequences; ribosomal RNA.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
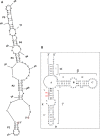
Fig. 1
An unusual 5S rRNA insertion. A schematic diagram showing (A) a possible secondary structure of the insert as predicted by mfold [50] and RNAstructure [51] and (B) the usual 5S rRNA secondary structure model (available at RNAcentral (
) and the Comparative RNA Web (CRW) Site(
http://www.rna.ccbb.utexas.edu
)) [52, 53] and modified; the insert is between positions U104 and G105 as per Halococcus morrhuae 5S rRNA numbering. The equivalent positions in Haloarcula marismortui are C108 and G109. What would be the 5′ and 3′ ends of the insert if it were an independent RNA are indicated in (A).
Fig. 2
Cryo‐EM reconstruction of the large subunit of Halococcus morrhuae. (A) The large subunit of Haloarcula marismotui (PDB:
1NJI
) [49] is rigidly docked in the density obtained from 3D cryo‐EM reconstruction; 5S rRNA (purple), 23S rRNA (yellow), and uL5 and uL18 (orange‐red), and selected ribosomal other proteins (cyan) are shown; the insert is marked by *; Cp—central protuberance. The scale bar indicates 50 Å. (B) A view of the large subunit with the best‐available crystal structure docked in (PDB:
4V9F
) [48] rotated to focus on the discordance in the helix 25 path (red arrow summarizing the crystal structure and green arrow summarizing the cryo‐EM structure). The cryo‐EM density at lower isosurface threshold is shown as an outline around the map, illustrating the continued projection of the helix. The P stalk can be seen on the left edge; though the P stalk proteins L11, L12, and L10e are visualized in the crystal structure of Haloarcula marismotui, they are not seen in this cryo‐EM map. Scale is identical to panel A. (C) Plot of the consistency of the two half‐maps after tight masking and mask correction. The 0.143 ‘gold‐standard’ cutoff is indicated as a dashed line.
Fig. 3
3D structure of the 5S rRNA. (A) The large subunit of Haloarcula marismotui (PDB:
1NJI
) [49] is docked in the density (left, view from the 30S side; right, view from the solvent side of the 50S subunit); only the 5S rRNA is shown colored, and the insert is marked by *. Coloring is by nucleotide: adenines, green; cytosines, orange; guanines, red; and uracils, blue. Cp—central protuberance; St—stalk. The scale bar indicates 50 Å. (B) The section of the map corresponding to the 5S rRNA is cropped out for visualization. The Haloarcula marismortui 5S rRNA was mutated to match the sequence of the Halococcus morrhuae 5S, except that the insert (whose secondary structure is not known) was omitted, and rigidly docked inside the density. The insertion site is marked by *. The scale bar indicates 5 Å. (C) The section of the map corresponding to the 5S rRNA is cropped out of the published, 6.6 Å Methanothermobacter thermautotrophicus cryo‐EM map and associated atomic model (EMD‐2012, PDB
4ADX
) [37] and colored as in (B). This and all other homolog maps lack the protruding lump of density seen in (B). Scale is identical to panel B.
Similar articles
- Very similar strains of Halococcus salifodinae are found in geographically separated permo-triassic salt deposits.
Stan-Lotter H, McGenity TJ, Legat A, Denner EBM, Glaser K, Stetter KO, Wanner G. Stan-Lotter H, et al. Microbiology (Reading). 1999 Dec;145 ( Pt 12):3565-3574. doi: 10.1099/00221287-145-12-3565. Microbiology (Reading). 1999. PMID: 10627054 - Halococcus qingdaonensis sp. nov., a halophilic archaeon isolated from a crude sea-salt sample.
Wang QF, Li W, Yang H, Liu YL, Cao HH, Dornmayr-Pfaffenhuemer M, Stan-Lotter H, Guo GQ. Wang QF, et al. Int J Syst Evol Microbiol. 2007 Mar;57(Pt 3):600-604. doi: 10.1099/ijs.0.64673-0. Int J Syst Evol Microbiol. 2007. PMID: 17329792 Free PMC article. - The 3D arrangement of the 23 S and 5 S rRNA in the Escherichia coli 50 S ribosomal subunit based on a cryo-electron microscopic reconstruction at 7.5 A resolution.
Mueller F, Sommer I, Baranov P, Matadeen R, Stoldt M, Wöhnert J, Görlach M, van Heel M, Brimacombe R. Mueller F, et al. J Mol Biol. 2000 Apr 21;298(1):35-59. doi: 10.1006/jmbi.2000.3635. J Mol Biol. 2000. PMID: 10756104 - Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy.
Greber BJ. Greber BJ. RNA. 2016 Nov;22(11):1643-1662. doi: 10.1261/rna.057927.116. RNA. 2016. PMID: 27875256 Free PMC article. Review. - Structure and function of 5S rRNA in the ribosome.
Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Bogdanov AA, et al. Biochem Cell Biol. 1995 Nov-Dec;73(11-12):869-76. doi: 10.1139/o95-094. Biochem Cell Biol. 1995. PMID: 8722002 Review.
Cited by
- Supersized Ribosomal RNA Expansion Segments in Asgard Archaea.
Penev PI, Fakhretaha-Aval S, Patel VJ, Cannone JJ, Gutell RR, Petrov AS, Williams LD, Glass JB. Penev PI, et al. Genome Biol Evol. 2020 Oct 1;12(10):1694-1710. doi: 10.1093/gbe/evaa170. Genome Biol Evol. 2020. PMID: 32785681 Free PMC article. - Expansion segments in bacterial and archaeal 5S ribosomal RNAs.
Stepanov VG, Fox GE. Stepanov VG, et al. RNA. 2021 Feb;27(2):133-150. doi: 10.1261/rna.077123.120. Epub 2020 Nov 12. RNA. 2021. PMID: 33184227 Free PMC article. - Ribosome Biogenesis in Archaea.
Londei P, Ferreira-Cerca S. Londei P, et al. Front Microbiol. 2021 Jul 22;12:686977. doi: 10.3389/fmicb.2021.686977. eCollection 2021. Front Microbiol. 2021. PMID: 34367089 Free PMC article. Review. - Looking through the Lens of the Ribosome Biogenesis Evolutionary History: Possible Implications for Archaeal Phylogeny and Eukaryogenesis.
Jüttner M, Ferreira-Cerca S. Jüttner M, et al. Mol Biol Evol. 2022 Apr 11;39(4):msac054. doi: 10.1093/molbev/msac054. Mol Biol Evol. 2022. PMID: 35275997 Free PMC article. - A Comparative Perspective on Ribosome Biogenesis: Unity and Diversity Across the Tree of Life.
Jüttner M, Ferreira-Cerca S. Jüttner M, et al. Methods Mol Biol. 2022;2533:3-22. doi: 10.1007/978-1-0716-2501-9_1. Methods Mol Biol. 2022. PMID: 35796979 Free PMC article. Review.
References
- Frank J and Agrawal RK (2000) A ratchet‐like inter‐subunit reorganization of the ribosome during translocation. Nature 406, 318–322. - PubMed
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M and Frank J (2003) Incorporation of aminoacyl‐tRNA into the ribosome as seen by cryo‐electron microscopy. Nat Struct Biol 10, 899–906. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources