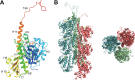Intracellular Organization by Jumbo Bacteriophages - PubMed (original) (raw)
Review
. 2020 Dec 18;203(2):e00362-20.
doi: 10.1128/JB.00362-20. Print 2020 Dec 18.
Affiliations
- PMID: 32868402
- PMCID: PMC7950403
- DOI: 10.1128/JB.00362-20
Review
Intracellular Organization by Jumbo Bacteriophages
Jingwen Guan et al. J Bacteriol. 2020.
Abstract
Since their discovery more than 100 years ago, the viruses that infect bacteria (bacteriophages) have been widely studied as model systems. Largely overlooked, however, have been "jumbo phages," with genome sizes ranging from 200 to 500 kbp. Jumbo phages generally have large virions with complex structures and a broad host spectrum. While the majority of jumbo phage genes are poorly functionally characterized, recent work has discovered many unique biological features, including a conserved tubulin homolog that coordinates a proteinaceous nucleus-like compartment that houses and segregates phage DNA. The tubulin spindle displays dynamic instability and centers the phage nucleus within the bacterial host during phage infection for optimal reproduction. The shell provides robust physical protection for the enclosed phage genomes against attack from DNA-targeting bacterial immune systems, thereby endowing jumbo phages with broad resistance. In this review, we focus on the current knowledge of the cytoskeletal elements and the specialized nuclear compartment derived from jumbo phages, and we highlight their importance in facilitating spatial and temporal organization over the viral life cycle. Additionally, we discuss the evolutionary relationships between jumbo phages and eukaryotic viruses, as well as the therapeutic potential and drawbacks of jumbo phages as antimicrobial agents in phage therapy.
Keywords: CRISPR-Cas immune systems; PhuZ tubulin filament; compartmentalization; jumbo bacteriophage; nucleus-like structure; phage therapy; protection; shell.
Copyright © 2020 Guan and Bondy-Denomy.
Figures
FIG 1
Monomeric and filament crystal structures of phage 201Ф2-1 PhuZ. (A) Cartoon representation of the crystal structure of a PhuZ201 monomer annotated with secondary structural elements (PDB ID
3R4V
) (53). The bound GDP-Mg2+ is shown in ball-and-stick format. (B) Cartoon representation of the crystal structure of PhuZ201 filament, with individual protofilaments presented in different colors (PDB ID
3J5V
) (67). The bound GDP-Mg2+ elements are shown in ball-and-stick format. (Left) Side view; (right) en d-on view.
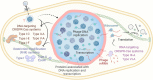
FIG 2
The bacteriophage nucleus-like shell combats antiphage defense mechanisms. Jumbo phages assemble a proteinaceous shell that separates phage genomes from the bacterial cytoplasm and segregates proteins according to function during viral replication. DNA replication and transcription occur inside the shell, while translation and metabolic processes take place in the cytoplasm. The shell physically shields phage genomes from attack by DNA-targeting CRISPR-Cas systems and restriction enzymes. However, RNA-targeting CRISPR-Cas systems can cleave phage transcripts in the cytoplasm. It remains unknown how phage DNA is protected upon injection (depicted as a dashed curve and a question mark).
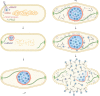
FIG 3
Intracellular development of jumbo phages after infection of a bacterial host. Upon encountering a bacterial host, the phage injects its genomic DNA, accompanied by the translocation of virion RNA polymerases (vRNAPs) that are present inside the viral capsid. vRNAPs transcribe early phage genes, including genes encoding the shell protein and PhuZ. While the host chromosome is degraded, PhuZ forms filaments anchored at each cell pole and gradually establishes a bipolar spindle in the cell. Meanwhile, the shell proteins assemble a nucleus-like compartment to sequester phage DNA from the cytoplasm and protect phage DNA against host defense systems. Nonvirion RNAPs (nvRNAPs) appear later inside the compartment and are responsible for expression of the late phage genes, including viral structural genes. As phage DNA replicates inside the shell, the compartment grows and is pushed toward the cell center by dynamically unstable filaments of the PhuZ spindle. The cell becomes elongated and forms a bulge at midcell. After the phage nucleus is settled in the cell center, treadmilling PhuZ filaments rotate the shell locally and deliver empty phage capsids from the cell inner membrane to the phage nucleus for subsequent DNA encapsidation. Rotation of the phage nucleus facilitates even distribution of phage capsids around the shell surface, ensuring efficient DNA packaging. Eventually, filled capsids are released from the shell and assemble into mature phage particles together with other viral structural components that are generated in the cytoplasm, followed by cell lysis to release phage progeny to the environment.
Similar articles
- The Phage Nucleus and PhuZ Spindle: Defining Features of the Subcellular Organization and Speciation of Nucleus-Forming Jumbo Phages.
Chaikeeratisak V, Birkholz EA, Pogliano J. Chaikeeratisak V, et al. Front Microbiol. 2021 Jul 13;12:641317. doi: 10.3389/fmicb.2021.641317. eCollection 2021. Front Microbiol. 2021. PMID: 34326818 Free PMC article. Review. - A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity.
Malone LM, Warring SL, Jackson SA, Warnecke C, Gardner PP, Gumy LF, Fineran PC. Malone LM, et al. Nat Microbiol. 2020 Jan;5(1):48-55. doi: 10.1038/s41564-019-0612-5. Epub 2019 Dec 9. Nat Microbiol. 2020. PMID: 31819217 - A cytoskeletal vortex drives phage nucleus rotation during jumbo phage replication in E. coli.
Birkholz EA, Laughlin TG, Armbruster E, Suslov S, Lee J, Wittmann J, Corbett KD, Villa E, Pogliano J. Birkholz EA, et al. Cell Rep. 2022 Aug 16;40(7):111179. doi: 10.1016/j.celrep.2022.111179. Cell Rep. 2022. PMID: 35977483 Free PMC article. - Type III CRISPR-Cas provides resistance against nucleus-forming jumbo phages via abortive infection.
Mayo-Muñoz D, Smith LM, Garcia-Doval C, Malone LM, Harding KR, Jackson SA, Hampton HG, Fagerlund RD, Gumy LF, Fineran PC. Mayo-Muñoz D, et al. Mol Cell. 2022 Dec 1;82(23):4471-4486.e9. doi: 10.1016/j.molcel.2022.10.028. Epub 2022 Nov 16. Mol Cell. 2022. PMID: 36395770 - Jumbo bacteriophages.
Hendrix RW. Hendrix RW. Curr Top Microbiol Immunol. 2009;328:229-40. doi: 10.1007/978-3-540-68618-7_7. Curr Top Microbiol Immunol. 2009. PMID: 19216440 Review.
Cited by
- Exploring pangenomic diversity and CRISPR-Cas evasion potential in jumbo phages: a comparative genomics study.
Magar S, Kolte V, Sharma G, Govindarajan S. Magar S, et al. Microbiol Spectr. 2024 Oct 3;12(10):e0420023. doi: 10.1128/spectrum.04200-23. Epub 2024 Sep 12. Microbiol Spectr. 2024. PMID: 39264185 Free PMC article. - Jumbo Phages: A Comparative Genomic Overview of Core Functions and Adaptions for Biological Conflicts.
M Iyer L, Anantharaman V, Krishnan A, Burroughs AM, Aravind L. M Iyer L, et al. Viruses. 2021 Jan 5;13(1):63. doi: 10.3390/v13010063. Viruses. 2021. PMID: 33466489 Free PMC article. - Bacteriophage genome engineering with CRISPR-Cas13a.
Guan J, Oromí-Bosch A, Mendoza SD, Karambelkar S, Berry JD, Bondy-Denomy J. Guan J, et al. Nat Microbiol. 2022 Dec;7(12):1956-1966. doi: 10.1038/s41564-022-01243-4. Epub 2022 Oct 31. Nat Microbiol. 2022. PMID: 36316452 Free PMC article. - Comparative genomics and proteomics analysis of phages infecting multi-drug resistant Escherichia coli O177 isolated from cattle faeces.
Montso PK, Kropinski AM, Mokoena F, Pierneef RE, Mlambo V, Ateba CN. Montso PK, et al. Sci Rep. 2023 Dec 5;13(1):21426. doi: 10.1038/s41598-023-48788-w. Sci Rep. 2023. PMID: 38052835 Free PMC article. - What makes a megaplasmid?
Hall JPJ, Botelho J, Cazares A, Baltrus DA. Hall JPJ, et al. Philos Trans R Soc Lond B Biol Sci. 2022 Jan 17;377(1842):20200472. doi: 10.1098/rstb.2020.0472. Epub 2021 Nov 29. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 34839707 Free PMC article.
References
- Summers WC. 2004. Bacteriophage research: early history, p 5–27. In Kutter E, Sulakvelidze A (ed), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources