A Simplified Quantitative Real-Time PCR Assay for Monitoring SARS-CoV-2 Growth in Cell Culture - PubMed (original) (raw)
. 2020 Sep 2;5(5):e00658-20.
doi: 10.1128/mSphere.00658-20.
Hung R Vuong # 1, Maritza Puray-Chavez 1, Adam L Bailey 2, Julie M Fox 3, Rita E Chen 2 3, Alex W Wessel 2 3, Jason M Scott 3, Houda H Harastani 3, Adrianus C M Boon 1 2 3, Haina Shin 3, Sebla B Kutluay 4
Affiliations
- PMID: 32878932
- PMCID: PMC7471006
- DOI: 10.1128/mSphere.00658-20
A Simplified Quantitative Real-Time PCR Assay for Monitoring SARS-CoV-2 Growth in Cell Culture
Christian Shema Mugisha et al. mSphere. 2020.
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected millions within just a few months, causing severe respiratory disease and mortality. Assays to monitor SARS-CoV-2 growth in vitro depend on time-consuming and costly RNA extraction steps, hampering progress in basic research and drug development efforts. Here, we developed a simplified quantitative real-time PCR assay that bypasses viral RNA extraction steps and can monitor SARS-CoV-2 growth from a small amount of cell culture supernatants. In addition, we show that this approach is easily adaptable to numerous other RNA and DNA viruses. Using this assay, we screened the activities of a number of compounds that were predicted to alter SARS-CoV-2 entry and replication as well as HIV-1-specific drugs in a proof-of-concept study. We found that E64D (inhibitor of endosomal proteases cathepsin B and L) and apilimod (endosomal trafficking inhibitor) potently decreased the amount of SARS-CoV-2 RNA in cell culture supernatants with minimal cytotoxicity. Surprisingly, we found that the macropinocytosis inhibitor ethylisopropylamiloride (EIPA) similarly decreased SARS-CoV-2 RNA levels in supernatants, suggesting that entry may additionally be mediated by an alternative pathway. HIV-1-specific inhibitors nevirapine (a nonnucleoside reverse transcriptase inhibitor [NNRTI]), amprenavir (a protease inhibitor), and allosteric integrase inhibitor 2 (ALLINI-2) modestly inhibited SARS-CoV-2 replication, albeit the 50% inhibitory concentration (IC50) values were much higher than that required for HIV-1. Taking the data together, this simplified assay will expedite basic SARS-CoV-2 research, be amenable to mid-throughput screening assays (i.e., drug, CRISPR, small interfering RNA [siRNA], etc.), and be applicable to a broad number of RNA and DNA viruses.IMPORTANCE Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of the coronavirus disease 2019 (COVID-19) pandemic, is continuing to cause immense respiratory disease and social and economic disruptions. Conventional assays that monitor SARS-CoV-2 growth in cell culture rely on costly and time-consuming RNA extraction procedures, hampering progress in basic SARS-CoV-2 research and development of effective therapeutics. Here, we developed a simple quantitative real-time PCR assay to monitor SARS-CoV-2 growth in cell culture supernatants that does not necessitate RNA extraction and that is as accurate and sensitive as existing methods. In a proof-of-concept screen, we found that E64D, apilimod, EIPA, and remdesivir can substantially impede SARS-Cov-2 replication, providing novel insight into viral entry and replication mechanisms. In addition, we show that this approach is easily adaptable to numerous other RNA and DNA viruses. This simplified assay will undoubtedly expedite basic SARS-CoV-2 and virology research and be amenable to use in drug screening platforms to identify therapeutics against SARS-CoV-2.
Keywords: SARS-CoV-2; antivirals; assay development; viral entry; virus replication.
Copyright © 2020 Shema Mugisha et al.
Figures
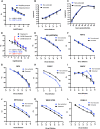
FIG 1
Development of a simplified qRT-PCR assay for SARS-CoV-2 viral RNA detection in cell culture supernatants. (A) Serially diluted RNA standards were either directly subjected to qRT-PCR or processed as described in the modified protocol detailed in the text prior to qRT-PCR. Log2 copy numbers are plotted against the cycle threshold (CT) values. Linear regression analysis was done to obtain the equations. Data show averages of results from three independent biological replicates. Error bars show standard errors of the means (SEM). (B) Comparison of the efficiency and detection ranges for quantifying SARS-CoV-2 RNA using purified RNA or lysed supernatants from virus stocks. Data are derived from three independent replicates. Error bars show the SEM. (C) Vero E6 cells were infected at a multiplicity of infection (MOI) of 0.01, and cell culture supernatants were analyzed for SARS-CoV-2 RNA following the conventional RNA extraction protocol versus the modified protocol developed here at various times postinfection. Cell-associated viral RNA was analyzed in parallel following RNA extraction for reference. Data are from three independent biological replicates. Error bars show the SEM. (D) Illustration of the efficiency and detection ranges of TaqMan-based and SYBR green-based qRT-PCR assays quantifying known amounts of SARS-CoV-2 RNA. Data are from 2 or 3 independent replicates. Error bars show the SEM. (E to L) The indicated viruses were subjected to RNA or DNA extraction (extracted) and diluted 10-fold or used directly following dilution (nonextracted) in the SYBR green-based (E to I) or TaqMan-based (J and L) qRT-PCR assay as described above. Samples were normalized such that equivalent amounts of the original virus stock were added to PCRs for extracted and nonextracted samples. Plots show the corresponding cycle threshold values (Ct, y axis) per virus dilution (x axis). Data are from two independent replicates, with error bars showing the SEM.
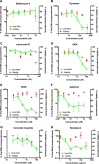
FIG 2
A compound screen to validate SARS-CoV-2-specific inhibitors and entry pathways. Vero E6 cells were infected with SARS-CoV-2 at an MOI of 0.01, and inhibitors were added concomitantly at the concentrations shown in the figures following virus adsorption. Supernatants from infected cells were lysed and used in a SYBR green-based qRT-PCR to quantify the viral RNA in cell culture supernatants. Compound cytotoxicity was monitored by the use of a RealTime-Glo MT cell viability assay kit (Promega) in parallel plates. Data show the cumulative results from 2 to 5 independent biological replicates. Error bars show the SEM. (A) Bafilomycin A. (B) Dynasore. (C) Latrunculin B. (D) EIPA. (E) E64D. (F) Apilimod. (G) Camostat mesylate. (H) Remdesivir.
Update of
- A facile Q-RT-PCR assay for monitoring SARS-CoV-2 growth in cell culture.
Mugisha CS, Vuong HR, Puray-Chavez M, Kutluay SB. Mugisha CS, et al. bioRxiv [Preprint]. 2020 Jun 28:2020.06.26.174698. doi: 10.1101/2020.06.26.174698. bioRxiv. 2020. PMID: 32607508 Free PMC article. Updated. Preprint.
Similar articles
- A facile Q-RT-PCR assay for monitoring SARS-CoV-2 growth in cell culture.
Mugisha CS, Vuong HR, Puray-Chavez M, Kutluay SB. Mugisha CS, et al. bioRxiv [Preprint]. 2020 Jun 28:2020.06.26.174698. doi: 10.1101/2020.06.26.174698. bioRxiv. 2020. PMID: 32607508 Free PMC article. Updated. Preprint. - Development of a Fluorescence-Based, High-Throughput SARS-CoV-2 3CLpro Reporter Assay.
Froggatt HM, Heaton BE, Heaton NS. Froggatt HM, et al. J Virol. 2020 Oct 27;94(22):e01265-20. doi: 10.1128/JVI.01265-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32843534 Free PMC article. - Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome.
Ye C, Chiem K, Park JG, Oladunni F, Platt RN 2nd, Anderson T, Almazan F, de la Torre JC, Martinez-Sobrido L. Ye C, et al. mBio. 2020 Sep 25;11(5):e02168-20. doi: 10.1128/mBio.02168-20. mBio. 2020. PMID: 32978313 Free PMC article. - Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2.
Li D, Zhang J, Li J. Li D, et al. Theranostics. 2020 Jun 1;10(16):7150-7162. doi: 10.7150/thno.47649. eCollection 2020. Theranostics. 2020. PMID: 32641984 Free PMC article. Review. - [Role of cyclophilin A during coronavirus replication and the antiviral activities of its inhibitors].
Tian L, Liu W, Sun L. Tian L, et al. Sheng Wu Gong Cheng Xue Bao. 2020 Apr 25;36(4):605-611. doi: 10.13345/j.cjb.200049. Sheng Wu Gong Cheng Xue Bao. 2020. PMID: 32347055 Review. Chinese.
Cited by
- Subversion of autophagy machinery and organelle-specific autophagy by SARS-CoV-2 and coronaviruses.
Sun Q, Li X, Kuang E. Sun Q, et al. Autophagy. 2023 Apr;19(4):1055-1069. doi: 10.1080/15548627.2022.2116677. Epub 2022 Aug 31. Autophagy. 2023. PMID: 36005882 Free PMC article. Review. - Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell.
Puray-Chavez M, LaPak KM, Schrank TP, Elliott JL, Bhatt DP, Agajanian MJ, Jasuja R, Lawson DQ, Davis K, Rothlauf PW, Jo H, Lee N, Tenneti K, Eschbach JE, Mugisha CS, Vuong HR, Bailey AL, Hayes DN, Whelan SPJ, Horani A, Brody SL, Goldfarb D, Major MB, Kutluay SB. Puray-Chavez M, et al. bioRxiv [Preprint]. 2021 Mar 1:2021.03.01.433431. doi: 10.1101/2021.03.01.433431. bioRxiv. 2021. PMID: 33688646 Free PMC article. Updated. Preprint. - Rilpivirine inhibits SARS-CoV-2 protein targets: A potential multi-target drug.
Ameen F, Mamidala E, Davella R, Vallala S. Ameen F, et al. J Infect Public Health. 2021 Oct;14(10):1454-1460. doi: 10.1016/j.jiph.2021.07.012. Epub 2021 Jul 21. J Infect Public Health. 2021. PMID: 34326009 Free PMC article. - Repurposing Approved Drugs for Guiding COVID-19 Prophylaxis: A Systematic Review.
Andrade BS, Rangel FS, Santos NO, Freitas ADS, Soares WRA, Siqueira S, Barh D, Góes-Neto A, Birbrair A, Azevedo VAC. Andrade BS, et al. Front Pharmacol. 2020 Dec 14;11:590598. doi: 10.3389/fphar.2020.590598. eCollection 2020. Front Pharmacol. 2020. PMID: 33390967 Free PMC article. Review. - A basally active cGAS-STING pathway limits SARS-CoV-2 replication in a subset of ACE2 positive airway cell models.
Puray-Chavez M, Eschbach JE, Xia M, LaPak KM, Zhou Q, Jasuja R, Pan J, Xu J, Zhou Z, Mohammed S, Wang Q, Lawson DQ, Djokic S, Hou G, Ding S, Brody SL, Major MB, Goldfarb D, Kutluay SB. Puray-Chavez M, et al. bioRxiv [Preprint]. 2024 Sep 6:2024.01.07.574522. doi: 10.1101/2024.01.07.574522. bioRxiv. 2024. PMID: 38260460 Free PMC article. Updated. Preprint.
References
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi:10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
- Chan JF, Yip CC, To KK, Tang TH, Wong SC, Leung KH, Fung AY, Ng AC, Zou Z, Tsoi HW, Choi GK, Tam AR, Cheng VC, Chan KH, Tsang OT, Yuen KY. 2020. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol 58:e00310-20. doi:10.1128/JCM.00310-20. - DOI - PMC - PubMed
- Sheahan TP, Sims AC, Leist SR, Schafer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO, Spahn JE, Bauer L, Sellers S, Porter D, Feng JY, Cihlar T, Jordan R, Denison MR, Baric RS. 2020. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun 11:222. doi:10.1038/s41467-019-13940-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI151810/AI/NIAID NIH HHS/United States
- U54 AI150470/AI/NIAID NIH HHS/United States
- DGE-1745038/National Science Foundation/International
- 1U01AI151810-01/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous