Cellular and Molecular Mechanisms of Metformin Action - PubMed (original) (raw)
Review
Cellular and Molecular Mechanisms of Metformin Action
Traci E LaMoia et al. Endocr Rev. 2021.
Abstract
Metformin is a first-line therapy for the treatment of type 2 diabetes, due to its robust glucose-lowering effects, well-established safety profile, and relatively low cost. While metformin has been shown to have pleotropic effects on glucose metabolism, there is a general consensus that the major glucose-lowering effect in patients with type 2 diabetes is mostly mediated through inhibition of hepatic gluconeogenesis. However, despite decades of research, the mechanism by which metformin inhibits this process is still highly debated. A key reason for these discrepant effects is likely due to the inconsistency in dosage of metformin across studies. Widely studied mechanisms of action, such as complex I inhibition leading to AMPK activation, have only been observed in the context of supra-pharmacological (>1 mM) metformin concentrations, which do not occur in the clinical setting. Thus, these mechanisms have been challenged in recent years and new mechanisms have been proposed. Based on the observation that metformin alters cellular redox balance, a redox-dependent mechanism of action has been described by several groups. Recent studies have shown that clinically relevant (50-100 μM) concentrations of metformin inhibit hepatic gluconeogenesis in a substrate-selective manner both in vitro and in vivo, supporting a redox-dependent mechanism of metformin action. Here, we review the current literature regarding metformin's cellular and molecular mechanisms of action.
Keywords: hepatic gluconeogenesis; metformin; redox; type 2 diabetes.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Endocrine Society.
Figures
Graphical Abstract
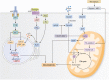
Figure 1.
Regulation of hepatic gluconeogenesis. Hepatic gluconeogenesis is regulated through allostery, substrate availability, redox balance, and gene expression. WAT lipolysis produces glycerol and NEFA, which can both independently stimulate hepatic gluconeogenesis. NEFA enters mitochondrial β-oxidation which produces acetyl-CoA, an allosteric activator of PC. PC catalyzes the conversion of pyruvate to oxaloacetate, which can directly enter the gluconeogenic pathway. On the other hand, glycerol is itself a gluconeogenic substrate. Glycerol from WAT lipolysis is phosphorylated to G3P and converted to DHAP, a gluconeogenic intermediate, by GPD2. The reaction catalyzed by GPD2 is dependent on cellular redox state and is inhibited by a high [NADH]:[NAD+] ratio. Similarly, gluconeogenesis from lactate is redox-regulated, because the conversion of lactate to pyruvate by LDH is inhibited by a high [NADH]:[NAD+] ratio. Transcriptional regulation of PCK1 and G6PC is coordinated by the opposing actions of glucagon and insulin. When insulin binds its receptor, AKT is activated and promotes nuclear exclusion of FOXO1, which decreases gluconeogenic gene expression. In contrast, glucagon binds the glucagon receptor and promotes IP3R-I-mediated ER Ca++ release. This in turn activates CRTC2, which forms a complex with CREB and CBP, and promotes transcriptional upregulation of PCK1 and G6PC. Abbreviations: CBP, CREB-binding protein; CREB, cAMP-responsive element-binding protein 1; CRTC2, CREB-regulated transcription co-activator 2; DHAP, dihydroxyacetone phosphate; DKI, double knock-in; FOXO, Forkhead box O; G3P, glycerol-3-phosphate; GCGR, glucagon receptor; INSR, insulin receptor; LDH, lactate dehydrogenase; NEFA, nonesterified fatty acid; PC, pyruvate carboxylase; WAT, white adipose tissue;
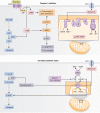
Figure 2.
Proposed mechanisms of metformin action. Complex I inhibition (top panel). Inhibition of complex I is central to several proposed mechanisms of metformin action. Following complex I inhibition, AMPK is activated by increased AMP levels, leading to inhibition of CRTC2 and preventing formation of the CREB-CBP-CRTC2 complex (orange boxes). AMPK also phosphorylates and inhibits ACC1 and 2, which promotes fat oxidation and decreases lipogenesis (purple boxes). Additionally, increased AMP is proposed to inhibit hepatic gluconeogenesis independently of AMPK. High AMP prevents glucagon-stimulated production of cAMP and therefore antagonizes hepatic glucagon action (pink boxes). AMP also allosterically inhibits FBP1, which directly inhibits the gluconeogenic pathway (blue boxes). Increased cytosolic redox (bottom panel). Metformin inhibition of GPD2 reduces the conversion of G3P to DHAP which impairs gluconeogenesis from glycerol and simultaneously increases the cytosolic [NADH]:[NAD+] ratio. LDH is inhibited by the increased [NADH]:[NAD+] ratio, thus reducing gluconeogenesis from lactate. Metformin inhibition of GPD2 is the only proposed mechanism that is independent of Complex I inhibition and produces substrate-selective inhibition of gluconeogenesis. Abbreviations: ACC, acetyl-CoA carboxylase; AMPK, adenosine monophosphate–activated protein kinase; CBP, CREB-binding protein; CREB, cAMP-responsive element-binding protein 1; CRTC2, CREB-regulated transcription co-activator 2; DHAP, dihydroxyacetone phosphate; FBP, fructose 1,6-bisphosphatase; FDP, fructose 1,6-diphosphate; G3P, glycerol-3-phosphate; GLP-1, glucagon-like peptide-1; GPD, glycerol-3-phosphate dehydrogenase; LDH, lactate dehydrogenase; OCT1, organic cation transporter 1.
References
- Pharmacologic approaches to glycemic treatment: “Standards of Medical Care in Diabetes—2020”. Diabetes Care. 2020;43:S98. - PubMed
- DeFronzo RA, Goodman AM. Efficacy of metformin in patients with non-insulin-dependent diabetes mellitus. N Engl J Med. 1995;333:541-549. - PubMed
- Hill J. The Vegetable System. London: R. Baldwin.
- Bailey CJ. Metformin: historical overview. Diabetologia. 2017;60(9):1566-1576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK113984/DK/NIDDK NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R01 AR080152/AR/NIAMS NIH HHS/United States
- T32 GM007324/GM/NIGMS NIH HHS/United States
- R01 DK114793/DK/NIDDK NIH HHS/United States
- R01 DK116774/DK/NIDDK NIH HHS/United States
- R01 DK119968/DK/NIDDK NIH HHS/United States
- RC2 DK120534/DK/NIDDK NIH HHS/United States
- P30 DK045735/DK/NIDDK NIH HHS/United States
- F31 DK126362/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical