Enterotoxigenic Escherichia coli infection promotes enteric defensin expression via FOXO6-METTL3-m6A-GPR161 signalling axis - PubMed (original) (raw)
Enterotoxigenic Escherichia coli infection promotes enteric defensin expression via FOXO6-METTL3-m6A-GPR161 signalling axis
Xin Zong et al. RNA Biol. 2021 Apr.
Abstract
The production of natural antimicrobial peptides has emerged as an important mechanism of innate immunity in animals. Defensins, members of a large family of antimicrobial peptides, have been suggested as effector molecules in host defence against bacteria, fungi, protozoa and enveloped viruses. However, the molecular mechanism underlying defensin upregulation in bacterial infection remains poorly understood. The modification of mRNA by N6-adenosine methylation (m6A) on internal bases influences gene expression in eukaryotes. Here, we show that β-defensin production triggered by Enterotoxigenic Escherichia coli K88 (E. coli K88) infection is controlled by the cellular m6A methyltransferase METTL3. Adding back with METTL3 robustly stimulated the re-expression of defensin, which further supports the conclusion. Furthermore, using a MeRIP-seq approach, we identified a functional connection between m6A dependent GPR161 signalling and the expression of defensins. Mechanistically, we found that the transcription factor FOXO6 interacted with METTL3 to trigger the transcription of GPR161 and the subsequent regulation of β-defensin expression. The study has shed light on the mechanisms by which enterotoxigenic Escherichia coli infection promotes enteric defensin expression.
Keywords: Enterotoxigenic Escherichia coli; FOXO6; GPR161; METTL3; N6-adenosine methylation; defensin.
Conflict of interest statement
No conflicts of interest exist.
Figures
Figure 1.
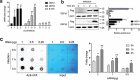
E. coli K88 simultaneously promotes the expression of β-defensin with m6A methylation. IPEC-J2 cells infected with E. coli K88 (MOI = 10:1) were analysed at various times, and the cells without infection constituted the control group. (A) The mRNA levels of DEFb1, DEFb2 and β-actin were measured by q-PCR, and the results are presented relative to those of Gapdh. (B) Immunoblotting was used to analyse the protein levels of DEFb1 and DEFb2 after transfection of the Flag fusion expression vector. The right panel shows the relative protein levels quantified by densitometry and normalized to the level of GFP. (C) m6A Dot blot was to measure the m6A levels with purified mRNA. Methylene blue staining was used as a loading control. The right panel shows the relative levels quantified by densitometry and normalized to the level of the control. The data are expressed as the mean ± SEM; statistically significant difference relative to the control: *P < 0.05, **P < 0.05, n = 3 biological replicates
Figure 2.
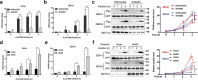
METTL3 depletion prevents β-defensin induction after E. coli K88 infection. IPEC-J2 cells with or without METTL3 knocking down were infected with E. coli K88 (MOI = 10:1) and analysed at various times. (A-B) The mRNA levels of DEFb1 (A) and DEFb2 (B) were measured by q-PCR, and the results are presented relative to the level of sh-Scramble 0 h group, and normalization to Gapdh. (C) Immunoblotting was used to analyse the protein levels of DEFb1 and DEFb2 after the transfection of the Flag fusion expression vector. The right panel shows the relative protein levels quantified by densitometry and normalized to the level of GFP. (D–F) METTL3-depleted IPEC-J2 cells were transfected with the Mettl3 plasmid or vector, followed by E. coli K88 infection and analysed at indicated times. The results from the q-PCR analysis of the mRNA levels of DEFb1 (D) and DEFb2 (E) are presented relative to the level of sh-Scramble 0 h group, and normalization to Gapdh. Immunoblotting was used to analyse the protein levels of Flag-labelled DEFb1 and DEFb2 (F). The right panel shows the relative protein levels quantified by densitometry and normalized to the level of GFP. The data are expressed as the mean ± SEM; *P < 0.05, **P < 0.05, n = 3 biological replicates
Figure 3.
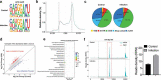
RNA m6A methylome profiles of IPEC-J2 cells after E. coli K88 infection. (A) Sequence motifs within m6A sites identified by using Homer software. (B) Pie chart depicting the m6A peaks as fractions. (C) Enrichment of m6A peaks in transcripts. Each transcript was divided into three parts: 5ʹUTR, CDS and 3ʹUTR. (D) The differentially m6A modified transcripts. (E) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of m6A differentially modified genes. (F) Peaks indicate the relative abundance of m6A sites in GPR161 mRNA. The m6A motif sequences that corresponded to an immunoprecipitate-enriched region are marked in green
Figure 4.
METTL3-mediated GPR161 is essential for β-defensin induction in intestinal epithelial cells. (A-B) METTL3-expressing and METTL3-knockdown IPEC-J2 cells were infected with E. coli K88 (MOI = 10:1) and analysed at various times. (A) The protein level of GPR161 was determined by immunoblot analysis. The right panel shows the relative protein levels quantified by densitometry and normalized to the level of β-actin. (B) Results of q-PCR used to quantify the mRNA level of GPR161 are presented relative to the level of sh-Scramble 0 h group, and normalization to Gapdh. (C-D) Transfected Mettl3 plasmid or vector into METTL3-depleted IPEC-J2 cells, followed by E. coli K88 infection for various times. Immunoblot analysis of the protein level of GPR161. The right panel shows the relative protein levels quantified by densitometry and normalized to the level of β-actin (C). Results from the q-PCR analysis of the mRNA level of GPR161 are presented relative to the level of sh-Scramble 0 h group, and normalization to Gapdh (D). (E) The mRNA levels of DEFb1 and DEFb2 in GPR161-expressing and GPR161-knockdown IPEC-J2 cells after E. coli K88 (MOI = 10:1) infection and analysed at various times. (F) The mRNA levels of DEFb1 and DEFb2 in GPR161-expressing and GPR161-overexpressing IPEC-J2 cells after E. coli K88 (MOI = 10:1) infection and analysed at various times. The data are expressed as the mean ± SEM; *P < 0.05, **P < 0.05, n = 3 biological replicates
Figure 5.
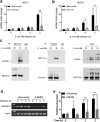
FOXO6 is involved in the correlation between METTL3 and GPR161. (A, B) Control of genome-wide responses to E. coli K88 by m6A methylase METTL3. (A) Volcano plots show differentially expressed genes (log2 fold change > 1) identified from RNA-seq of polyadenylated RNA collected from cells with or without METTL3. (B) Path-way analyses (KEGG) of significantly differentially expressed genes from (a) were conducted using DAVID and filtered according to a Benjamini-Hochberg procedure (<0.05). (C, D) IPEC-J2 cells with or without METTL3 knockdown were infected with E. coli K88 (MOI = 10:1) for indicated times. Immunoblot analysis of FOXO6 protein levels. The right panel shows the relative protein levels quantified by densitometry and normalized to β-actin (C). q-PCR analysis of the mRNA level of FOXO6, and the results are presented relative to the level of sh-Scramble 0 h group, and normalization to Gapdh(D). (E-F) Transfected Mettl3 plasmid or vector into METTL3-depleted IPEC-J2 cells, followed by E. coli K88 infection for various times. (E) Immunoblot analysis of FOXO6 protein levels. The right panel shows the relative protein levels quantified by densitometry and normalized to the level of β-actin. (F) Results from the q-PCR analysis of the mRNA level of FOXO6 are presented relative to the level of sh-Scramble 0 h group, and normalization to Gapdh. (G) The mRNA level of GPR161 in FOXO6-expressing and FOXO6-knockdown IPEC-J2 cells after E. coli K88 (MOI = 10:1) infection and analysed for indicated times. The data are expressed as the mean ± SEM; *P < 0.05, **P < 0.05, n = 3 biological replicates
Figure 6.
FOXO6 interacts with METTL3 to initiate the transcription of GPR161. (A, B) The mRNA levels of DEFb1 (A) and DEFb2 (B) in FOXO6-expressing and FOXO6-knockdown IPEC-J2 cells after E. coli K88 (MOI = 10:1) infection and analysed at indicated times. (C, –D) Immunoprecipitation was performed with antibodies against METTL3 (C) or FOXO6 (D), with immunoglobulin G (IgG) serving as the control. The samples were analysed by immunoblot assay using the indicated antibodies. (E–F) ChIP analysis was performed using an FOXO6 antibody (E) and q-PCR (F). The data are expressed as the mean ± SEM; *P < 0.05, **P < 0.05, n = 3 biological replicates
Similar articles
- Protective effects of Lactobacillus plantarum on epithelial barrier disruption caused by enterotoxigenic Escherichia coli in intestinal porcine epithelial cells.
Wu Y, Zhu C, Chen Z, Chen Z, Zhang W, Ma X, Wang L, Yang X, Jiang Z. Wu Y, et al. Vet Immunol Immunopathol. 2016 Apr;172:55-63. doi: 10.1016/j.vetimm.2016.03.005. Epub 2016 Mar 5. Vet Immunol Immunopathol. 2016. PMID: 27032504 - Dietary supplementation with Clostridium butyricum helps to improve the intestinal barrier function of weaned piglets challenged with enterotoxigenic Escherichia coli K88.
Li HH, Li YP, Zhu Q, Qiao JY, Wang WJ. Li HH, et al. J Appl Microbiol. 2018 Oct;125(4):964-975. doi: 10.1111/jam.13936. Epub 2018 Jul 17. J Appl Microbiol. 2018. PMID: 29851202 - Protective effect of chicken egg yolk immunoglobulins (IgY) against enterotoxigenic Escherichia coli K88 adhesion in weaned piglets.
Wang Z, Li J, Li J, Li Y, Wang L, Wang Q, Fang L, Ding X, Huang P, Yin J, Yin Y, Yang H. Wang Z, et al. BMC Vet Res. 2019 Jul 8;15(1):234. doi: 10.1186/s12917-019-1958-x. BMC Vet Res. 2019. PMID: 31286936 Free PMC article. - Host-microbe interaction: mechanisms of defensin deficiency in Crohn's disease.
Wang G, Stange EF, Wehkamp J. Wang G, et al. Expert Rev Anti Infect Ther. 2007 Dec;5(6):1049-57. doi: 10.1586/14787210.5.6.1049. Expert Rev Anti Infect Ther. 2007. PMID: 18039087 Review. - Intestinal receptors for adhesive fimbriae of enterotoxigenic Escherichia coli (ETEC) K88 in swine--a review.
Jin LZ, Zhao X. Jin LZ, et al. Appl Microbiol Biotechnol. 2000 Sep;54(3):311-8. doi: 10.1007/s002530000404. Appl Microbiol Biotechnol. 2000. PMID: 11030565 Review.
Cited by
- TRIM28 recruits E2F1 to regulate CBX8-mediated cell proliferation and tumor metastasis of ovarian cancer.
Zhang F, Zhu T, Wu C, Shen D, Liu L, Chen X, Guan Y, Ding H, Tong X. Zhang F, et al. Hum Cell. 2023 Nov;36(6):2113-2128. doi: 10.1007/s13577-023-00983-7. Epub 2023 Sep 14. Hum Cell. 2023. PMID: 37709991 - Multilevel regulation of N6-methyladenosine RNA modifications: Implications in tumorigenesis and therapeutic opportunities.
Feng L, Du R, Chang B, Li M, Tian J, Wang S. Feng L, et al. Genes Dis. 2022 Sep 7;10(5):1969-1981. doi: 10.1016/j.gendis.2022.08.018. eCollection 2023 Sep. Genes Dis. 2022. PMID: 37492716 Free PMC article. Review. - The evolving landscape of N6-methyladenosine modification in the tumor microenvironment.
Gu Y, Wu X, Zhang J, Fang Y, Pan Y, Shu Y, Ma P. Gu Y, et al. Mol Ther. 2021 May 5;29(5):1703-1715. doi: 10.1016/j.ymthe.2021.04.009. Epub 2021 Apr 9. Mol Ther. 2021. PMID: 33839323 Free PMC article. Review. - Mechanisms and regulation of defensins in host defense.
Fu J, Zong X, Jin M, Min J, Wang F, Wang Y. Fu J, et al. Signal Transduct Target Ther. 2023 Aug 14;8(1):300. doi: 10.1038/s41392-023-01553-x. Signal Transduct Target Ther. 2023. PMID: 37574471 Free PMC article. Review. - Intracellular Fusobacterium nucleatum infection increases METTL3-mediated m6A methylation to promote the metastasis of esophageal squamous cell carcinoma.
Guo S, Chen F, Li L, Dou S, Li Q, Huang Y, Li Z, Liu W, Zhang G. Guo S, et al. J Adv Res. 2024 Jul;61:165-178. doi: 10.1016/j.jare.2023.08.014. Epub 2023 Aug 22. J Adv Res. 2024. PMID: 37619934 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
This work was funded by National Natural Science Foundation of China [grants no 31601947.
LinkOut - more resources
Full Text Sources
Other Literature Sources