Joint analysis of lncRNA m6A methylome and lncRNA/mRNA expression profiles in gastric cancer - PubMed (original) (raw)
Joint analysis of lncRNA m6A methylome and lncRNA/mRNA expression profiles in gastric cancer
Zhi Lv et al. Cancer Cell Int. 2020.
Abstract
Background: N 6-methyladenosine (m6A) modification might be closely associated with the genesis and development of gastric cancer (GC). Currently, the evidence established by high-throughput assay for GC-related m6A patterns based on long non-coding RNAs (lncRNAs) remains limited. Here, a joint analysis of lncRNA m6A methylome and lncRNA/mRNA expression profiles in GC was performed to explore the regulatory roles of m6A modification in lncRNAs.
Methods: Three subjects with primary GC were enrolled in our study and paired sample was randomly selected from GC tissue and adjacent normal tissue for each case. Methylated RNA Immunoprecipitation NextGeneration Sequencing (MeRIP-Seq) and Microarray Gene Expression Profiling was subsequently performed. Then co-expression analysis and gene enrichment analysis were successively conducted.
Results: After data analysis, we identified 191 differentially m6A-methylated lncRNAs, 240 differentially expressed lncRNAs and 229 differentially expressed mRNAs in GC. Furthermore, four differentially m6A-methylated and expressed lncRNAs (dme-lncRNAs) were discovered including RASAL2-AS1, LINC00910, SNHG7 and LINC01105. Their potential target genes were explored by co-expression analysis. And gene enrichment analysis suggested that they might influence the cellular processes and biological behaviors involved in mitosis and cell cycle. The potential impacts of these targets on GC cells were further validated by CCLE database and literature review.
Conclusions: Four novel dme-lncRNAs were identified in GC, which might exert regulatory roles on GC cell proliferation. The present study would provide clues for the lncRNA m6A methylation-based research on GC epigenetic etiology and pathogenesis.
Keywords: Expression profile; Gastric cancer; LncRNA; Methylome profile; m6A.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures
Fig. 1
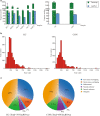
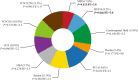
The overall features of lncRNA m6A methylome in GC. a the quantitative data of RNA transcripts in MeRIP and m6A-methylated lncRNAs; b the peak width distribution of lncRNA m6A-methylated sites in sequencing; c the source distribution of m6A-methylated lncRNAs
Fig. 2
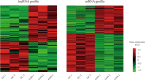
The heat map of expression levels for differentially expressed lncRNAs and mRNAs in GC
Fig. 3
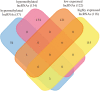
The Venn diagram of differentially m6A-methylated lncRNAs and differentially expressed lncRNAs in GC
Fig. 4
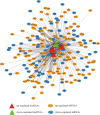
The co-expression network of dme-lncRNAs and differentially co-expressed mRNAs in GC
Fig. 5
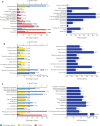
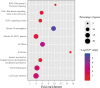
The prediction of top 10 expression sites in the enrichment analysis of differentially co-expressed genes in GC. FE fold enrichment
Fig. 6
The GO-term enrichment analysis of differentially co-expressed genes in GC. Top 10 items were selected for each term. a cellular component; b molecular function; c biological process
Fig. 7
The pathway analysis of differentially co-expressed genes in GC. Top 10 items were selected
Fig. 8
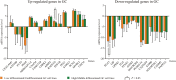
The differences in mRNA expression levels of selected differentially co-expressed genes in GC cell lines grouped by differentiation
Similar articles
- Depicting the Profile of METTL3-Mediated lncRNA m6A Modification Variants and Identified SNHG7 as a Prognostic Indicator of MNNG-Induced Gastric Cancer.
Liu T, Feng Y, Yang S, Ge Y, Zhang T, Li J, Li C, Ruan Y, Luo B, Liang G. Liu T, et al. Toxics. 2023 Nov 20;11(11):944. doi: 10.3390/toxics11110944. Toxics. 2023. PMID: 37999596 Free PMC article. - High-Throughput Sequencing Reveals N6-Methyladenosine-modified LncRNAs as Potential Biomarkers in Mice with Liver Fibrosis.
Wu F, Zhang S, Fan C, Huang S, Jiang H, Zhang J. Wu F, et al. Curr Gene Ther. 2023;23(5):371-390. doi: 10.2174/1566523223666230606151013. Curr Gene Ther. 2023. PMID: 37282641 - Integrated analysis of long non-coding RNAs and mRNA expression profiles reveals the potential role of lncRNAs in gastric cancer pathogenesis.
Lin XC, Zhu Y, Chen WB, Lin LW, Chen DH, Huang JR, Pan K, Lin Y, Wu BT, Dai Y, Tu ZG. Lin XC, et al. Int J Oncol. 2014 Aug;45(2):619-28. doi: 10.3892/ijo.2014.2431. Epub 2014 May 9. Int J Oncol. 2014. PMID: 24819045 - Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.
Luo R, Song J, Zhang W, Ran L. Luo R, et al. Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review. - New insights on the interplays between m6A modifications and microRNA or lncRNA in gastrointestinal cancers.
Su T, Liu J, Zhang N, Wang T, Han L, Wang S, Yang M. Su T, et al. Front Cell Dev Biol. 2023 Jun 19;11:1157797. doi: 10.3389/fcell.2023.1157797. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37404673 Free PMC article. Review.
Cited by
- RNA m6 A methylation in cancer.
Wang Z, Zhou J, Zhang H, Ge L, Li J, Wang H. Wang Z, et al. Mol Oncol. 2023 Feb;17(2):195-229. doi: 10.1002/1878-0261.13326. Epub 2022 Nov 6. Mol Oncol. 2023. PMID: 36260366 Free PMC article. Review. - Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis.
Zhou Y, Jiang R, Jiang Y, Fu Y, Manafhan Y, Zhu J, Jia E. Zhou Y, et al. Cells. 2022 Sep 24;11(19):2980. doi: 10.3390/cells11192980. Cells. 2022. PMID: 36230944 Free PMC article. - LncRNA RASAL2-AS1 promotes METTL14-mediated m6A methylation in the proliferation and progression of head and neck squamous cell carcinoma.
Rong M, Zhang M, Dong F, Wu K, Cai B, Niu J, Yang L, Li Z, Lu HY. Rong M, et al. Cancer Cell Int. 2024 Mar 25;24(1):113. doi: 10.1186/s12935-024-03302-8. Cancer Cell Int. 2024. PMID: 38528591 Free PMC article. - The regulatory mechanism of m6A modification in gastric cancer.
Wu S, Li C, Zhou H, Yang Y, Liang N, Fu Y, Luo Q, Zhan Y. Wu S, et al. Discov Oncol. 2024 Jul 15;15(1):283. doi: 10.1007/s12672-024-00994-2. Discov Oncol. 2024. PMID: 39009956 Free PMC article. Review. - RNA N6-methyladenosine methylation in influenza A virus infection.
Liu X, Chen W, Li K, Sheng J. Liu X, et al. Front Microbiol. 2024 Jun 18;15:1401997. doi: 10.3389/fmicb.2024.1401997. eCollection 2024. Front Microbiol. 2024. PMID: 38957616 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous