Negative feedback control of neuronal activity by microglia - PubMed (original) (raw)
. 2020 Oct;586(7829):417-423.
doi: 10.1038/s41586-020-2777-8. Epub 2020 Sep 30.
Ana Badimon 1 2 3, Hayley J Strasburger 1 2 3, Pinar Ayata 1 2 3 4, Aditya Nair 5, Ako Ikegami 6 7, Philip Hwang 1 2 3, Andrew T Chan 1 2 3, Steven M Graves 8, Joseph O Uweru 9, Carola Ledderose 10, Munir Gunes Kutlu 11, Michael A Wheeler 12, Anat Kahan 5, Masago Ishikawa 1, Ying-Chih Wang 13, Yong-Hwee E Loh 1, Jean X Jiang 14, D James Surmeier 15, Simon C Robson 16 17, Wolfgang G Junger 10, Robert Sebra 13, Erin S Calipari 11 18 19 20 21, Paul J Kenny 1, Ukpong B Eyo 9, Marco Colonna 22, Francisco J Quintana 12 23, Hiroaki Wake 6 7, Viviana Gradinaru 5, Anne Schaefer 24 25 26 27
Affiliations
- PMID: 32999463
- PMCID: PMC7577179
- DOI: 10.1038/s41586-020-2777-8
Negative feedback control of neuronal activity by microglia
Ana Badimon et al. Nature. 2020 Oct.
Abstract
Microglia, the brain's resident macrophages, help to regulate brain function by removing dying neurons, pruning non-functional synapses, and producing ligands that support neuronal survival1. Here we show that microglia are also critical modulators of neuronal activity and associated behavioural responses in mice. Microglia respond to neuronal activation by suppressing neuronal activity, and ablation of microglia amplifies and synchronizes the activity of neurons, leading to seizures. Suppression of neuronal activation by microglia occurs in a highly region-specific fashion and depends on the ability of microglia to sense and catabolize extracellular ATP, which is released upon neuronal activation by neurons and astrocytes. ATP triggers the recruitment of microglial protrusions and is converted by the microglial ATP/ADP hydrolysing ectoenzyme CD39 into AMP; AMP is then converted into adenosine by CD73, which is expressed on microglia as well as other brain cells. Microglial sensing of ATP, the ensuing microglia-dependent production of adenosine, and the adenosine-mediated suppression of neuronal responses via the adenosine receptor A1R are essential for the regulation of neuronal activity and animal behaviour. Our findings suggest that this microglia-driven negative feedback mechanism operates similarly to inhibitory neurons and is essential for protecting the brain from excessive activation in health and disease.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures
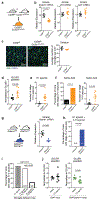
Extended Data Figure 1.. DREADD-based mouse models to study microglia responses to neuronal activation and inhibition reveals distinct microglia responses.
a, b, Neuron-specific activation (a) and inhibition (b) has been achieved by the expression of the Gq-coupled (activating) hM3Dq or Gi-coupled (inhibiting) hM4Di in CaMKII+ forebrain neurons. The CaMKII-tTa mice were bred to either tetO-CHRM3 or tetO-CHRM4 mice to generate CaMKII-tTa;tetO-CHRM3 or CaMKII-tTa;tetO-CHRM4 mice_._ hM3Dq or hM4Di were activated by i.p. injection of clozapine-N-oxide (CNO) to activate (0.25mg/kg) or inhibit (1mg/kg) CaMKII+ neuronal activity, respectively. c-e, Validation of CNO-mediated neuronal activation and inhibition: c, Heatmap (left) and violin plot (right) show RNA expression levels of 18 immediate early genes in total striatum 2 hours after CNO-mediated neuronal inhibition (orange) or neuronal activation (blue) as compared with controls (n=2 CaMKII-tTa;tetO-CHRM4, n=5 control, and n=3 CaMKII-tTa;tetO-CHRM3 mice) (right, _P_=0.0001, One-way ANOVA (Kruskal–Wallis test) with Dunn’s multiple comparison test). d, Dot plot showing quantification of the average number of cFOS+ cells in the dorsal striatum of CaMKII-tTa;tetO-CHRM4 (orange, n=4 mice), control (black, n=6 mice), and CaMKII-tTa;tetO-CHRM3 (blue, n=4 mice) mice one hour after treatment with CNO (_P_=0.0004, One-way ANOVA with Tukey’s post hoc test). e, Representative images showing cFOS+ cells (green) in the striatum of CaMKII-tTa;tetO-CHRM4 (top), control (middle), and CaMKII-tTa;tetO-CHRM3 (bottom) mice in response to CNO, DAPI (blue) (image are representative of two independent cohorts of mice). f, To allow for the microglia-specific analysis of changes in ribosome-associated RNA levels following neuron inhibition, the CaMKII-tTa;tetO-CHRM4 mice were bred to Cx3cr1CreErt2/+(Litt); Eef1a1LSL.eGFPL10a/+ mice followed by tamoxifen-induced Cre-mediated L10a-eGFP expression in microglia. g, Changes in ribosome-bound mRNA levels in striatal microglia were determined using the TRAP-sequencing approach. The heatmap shows the variation in the expression levels of 135 upregulated and 220 downregulated genes (_z_-scored log2(RPKM) at 2 hours following CNO-mediated neuronal inhibition. h, Selected gene ontology (GO) annotations for upregulated genes (DESeq2) in striatal microglia in response to neuronal inhibition, GO analysis was performed using ENRICHR analysis, (dotted line, _P_=0.05). i, Venn diagrams comparing microglial genes up- and down-regulated following CaMKII + neuronal activation and inhibition reveals highly differential microglia response. j, qPCR confirmation of increased mRNA expression (lower ΔCT, normalized to Gapdh) in microglia upon neuronal activation (Ccl3, left, n=3 mice, _P_=0.059, unpaired two-tailed t-test) and neuronal inhibition (Cd74, right, n=2 mice). k, Dot plots show lack of expression changes in selected genes in the striatum of wild type mice 2 hrs after saline, 0.25mg/kg CNO injection, or 1mg/kg CNO injection (n=3, 3, and 3 mice; _Kdm6b: P_=0.70 _Adrb1: P_=0.22, _Ccl24: P_=0.54, _Ccl3: P_=0.43, _Kcnk13: P_=0.37, _Ikbkb, P_=0.62, One-way ANOVA with Tukey’s post hoc test). RPKM: reads per kilobase of transcript per million mapped reads, TRAP: translating ribosome affinity purification; Data shown as mean± s.e.m.
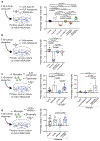
Extended Data Figure 2.. Microglia deficient mice show normal baseline behaviors but exaggerated responses to neurostimulants.
a, Dot plots show the average number of microglia per mm in cortex, striatum, cerebellum and hippocampus in control and microglia deficient mice (n=3 and 4 mice, cortex: P<0.0001, striatum: _P_=0.0003, cerebellum: _P_=0.0001, DG: P<0.0001, CA3: P<0.0001, CA1: P<0.0001, unpaired two-tailed t-test). b-e, Behavioral characteristics of microglia deficient mice**. b,** Anxiety-like behavior was measured by the ratio of time spent in the open arms/closed arms in the elevated plus maze (n=10 mice, _P_=0.65, unpaired two-tailed t-test). c, Motor coordination was measured by latency to fall from the accelerating rotarod (n=8 and12 mice, interaction: _P_=0.89, time: _P_=0.13, treatment: _P_=0.36; subjects: P<0.0001, Two-way repeated measures ANOVA). d, Olfactory behavior was measured by the sniff test (n=21 and 13 mice, _P_=0.09, unpaired two-tailed t-test). e, Social behavior was measured by using the classic three-chamber sociability task (Social preference: mouse preference for sniffing another mouse over object, Control: n=7 mice; _P_=0.0002, microglia deficient: n=9 mice, P<0.0001; Social Memory: mouse preference for sniffing novel mouse over familiar mouse, Control n=7 mice, _P_=0.0023, microglia deficient: n=7 mice, _P_=0.0009; paired two-tailed t-test). f, Representative images show brain-wide gene expression patterns of receptors targeted by kainic acid (kainate and AMPA receptor), picrotoxin (GABAA receptor), and SKF81297 (D1 receptor) (Allen Institute). g, Number of stage IV-V seizures (Racine scale) per mouse visually recorded within one hour in response to kainic acid (18mg/kg, i.p.) are shown as a dot plot (n=9 and 10 mice, _P_=0.0008, unpaired two-tailed t-test). h, Dot plot showing distance traveled in response to D1 agonist in one hour in the open field (SKF81297, 3mg/kg, i.p.)(n=14 and 8 mice, _P_=0.025, unpaired two-tailed t-test). i, Representative cortical EEG traces during a tonic-clonic seizure event in response to D1 agonist treatment (SKF81297, 5mg/kg i.p.) in control (top) and microglia deficient (bottom) mice showing high amplitude and rhythmic discharges followed by EEG depression. DG: dentate gyrus; Data shown as mean± s.e.m.
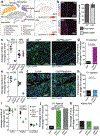
Extended Data Figure 3.. Generation and characterization of _Il34_-deficient and _Csf1_-deficient mice.
a, Violin plots show the expression levels of cell-type specific representative marker genes across the 10 identified cell types from striatum single nuclei RNA-seq (snRNA-seq) data analysis. Black dots indicate mean expression of selected gene per cell type. b, In situ hybridization for Il34 (left) and Csf1 (right) mRNAs show differential, region-specific expression in cortex, striatum, CA1, dentate gyrus (DG), CA3, corpus callosum (CC), and cerebellum of wild-type mice (WM: white matter, GM: grey matter, ML: molecular layer, GCL: granule cell layer, scale bar=100μm). c, h, The striatal grey matter-specific or white matter-specific microglia depletion was achieved by breeding NestinCre/+ mice to Il34fl/fl mice or Csf1fl/fl mice, respectively, to generate Il34fl/fl;NestinCre/+ (purple, c) and Csf1fl/fl;NestinCre/+ mice (blue, h). d, i, Dot plots showing relative expression levels of Il34 and Csf1 mRNA normalized to Gapdh in the striatum of Il34fl/fl;NestinCre/+ mice (d) or Csf1fl/fl;NestinCre/+ mice (i) compared with littermate controls (d, n=4 mice each, Il34 P<0.0001, Csf1 P =0.69; i, n=3 and 5 mice, _Il34 P_=0.07, Csf1 p<0.0001, unpaired two-tailed t-test). e, Dot plots show the average microglia density per mm per mouse in cortex, striatum, cerebellum (cortex: n=9, 12, and 10 mice, P<0.0001, striatum: n=9, 13, and 10 mice, P<0.0001, cerebellum: n=7, 7, and 8 mice, _P_=0.34, One-way ANOVA with Tukey’s post hoc test). f, left, Dot plot shows levels of IL34 protein as determined by Western blot analysis of striatal protein lysate from Il34fl/fl, Il34fl/+;NestinCre/+ or Il34fl/fl;NestinCre/+ mice normalized to DARPP32 expression (n=3 mice, _P_=0.0077, One-way ANOVA with Tukey’s post hoc test). g, j, Bar graphs show the average percentage of white matter regions in striatal images (0.5mm x 0.5mm) used to count WM and GM microglia in control and mutant mice for the data shown in Fig. 2c and e. (g, P = 0.99, n = 4 and 3 mice, unpaired two-tailed t-test; j, n = 4 and 2 a mice). For gel source data, see Supplementary Figure 1. Data shown as mean± s.e.m.
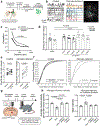
Extended Data Figure 4.. Generation of mice with striatum-specific microglia depletion.
a, b, (left), The striatum-specific microglia depletion was achieved by breeding Il34fl/fl mice to Drd1aCre/+ or Drd2Cre/+ mice to generate Il34fl/fl;Drd1aCre/+ (a, green) and Il34fl/fl;Drd2Cre/+ mice (b, grey). Right, dot plots show relative expression of Il34 mRNA in the striatum normalized to Gapdh (a, n=6 and 7 mice, P<0.0001; b, n=4 mice, _P_=0.0004, unpaired two-tailed t-test). c, Representative striatal images of sagittal brain slices from Il34fl/fl, Il34fl/fl;Drd1aCre/+ and Il34fl/fl;Drd2Cre/+ mice following immunofluorescent staining for P2RY12 (microglia, green) and DAPI (nuclei, blue) (scale bar = 50μm). d, e, Dot plots show the average microglia density per mm per mouse per specific region in the hippocampus of Il34fl/fl;Drd1aCre/+ (d) and Il34fl/fl;Drd2Cre/+ mice (e) compared to littermate controls (d, n=3 mice, DG: _P_=0.88, CA3: _P_=0.85, CA1: _P_=0.1; e, n=3 mice, DG: _P_=0.69, CA3: _P_=0.56, CA1: _P_=0.72; unpaired two-tailed t-test). f, g, Dot plots showing total distance traveled in response to D1 agonist (SKF81297, 3mg/kg, i.p.) in one hour in the open field for Il34fl/fl;Drd1aCre/+ (f) and Il34fl/fl;Drd2Cre/+ mice (g) compared with littermate controls (f: n=8 and 9 mice, _P_=0.034 g: n=8 mice, _P_=0.0087, unpaired two-tailed t-test). h, Percentage of mice seizing 30 minutes after administration of picrotoxin (i.p., 1mg/kg) shown as a bar graph (n=21, 9, and 8 mice; _P_=0.80, Chi-squared test). DG: dentate gyrus. i, Microglia-neuron ratio defines the threshold of D1 neuron activation by D1 agonist. Bar graph shows the percentage of mice with stage IV-V seizures in response to D1 agonist (4mg/kg, i.p.) in control, Il34fl/fl;Drd1aCre/+, and microglia deficient mice (n=11, 13, and 9 mice; right, _P_=0.0005, Chi-squared test). While all mice display an increased seizure response to 5mg/kg D1 agonist treatment, only microglia deficient (99% reduction of microglia), but not Il34fl/fl;Drd1aCre/+ (60% reduction of microglia in the striatum) display an increased seizure response at 4mg/kg D1 agonist treatment. Data shown as mean± s.e.m.
Extended Data Figure 5.. Striatum-specific microglia reduction has no overall effects on striatal cellular composition, D1/D2 neuronal morphology, D1/D2 MSN characteristic electrophysiological and molecular phenotypes, and glial phenotypes.
a, Dot plots show average number of D1 neurons (left, dark green, GFP+, DARPP32+) and D2 neurons (right, light green, GFP-, DARPP32+) per mouse in the striatum of Il34fl/fl;Drd1aeGFPL10a and Il34fl/fl;Drd1aeGFPL10a;Drd1aCre/+ mice. Mice expressing eGFP-tagged ribosomal subunit L10a under the Drd1a promoter were used to identify GFP+ D1 neurons and GFP− D2 neurons in control Il34fl/fl;Drd1aeGFPL10a and mutant Il34fl/fl;Drd1aeGFPL10a;Drd1aCre/+ (n=2 mice). b, c, D1 or D2 neuron cell morphology was determined by the number of primary dendrites (b), total dendritic length (c, left), and sholl analysis (c, right) (b, D1 neurons: n=11 and 15 D1 neurons, _P_=0.33; D2 neurons: n=15 and 11 D2 neurons, _P_=0.59; unpaired two-tailed t-test; c, D1 neurons, n=11 and 15 D1 neurons, dendritic length: _P_=0.83, unpaired two-tailed t-test; sholl, interaction: _P_=0.99; genotype: _P_=0.069; distance: P<0.0001, two-way ANOVA; D2 neurons, n=15 and 10 D2 neurons, dendritic length: _P_=0.80, unpaired two-tailed t-test; sholl: interaction: _P_=0.051; genotype: _P_=0.67; distance: _P_<0.0001, two-way ANOVA). **d,** Intrinsic excitability of D1 neurons (left) and D2 neurons (right) in _ex vivo_ slices as measured by current-evoked action potentials (AP, left) and equilibrium potentials as voltage-current (VC) plots (right) (D1**:** n=11 and 15 D1 neurons, AP: interaction: _P_=1.0; genotype: _P_=0.98; pA: _P_<0.0001, subjects: _P_<0.0001; VC: interaction: _P_=1.0; genotype: _P_=0.48; distance: _P_<0.0001, subjects: _P_<0.0001; D2: n=16 and 10 D2 neurons; AP: interaction: _P_=1.0; genotype: _P_=0.5; distance: _P_<0.0001, subjects: _P_<0.0001; VC: interaction: _P_=0.99; genotype: _P_=0.7; distance: _P_<0.0001, subjects: _P_<0.0001; two-way ANOVA). **e,** Dendritic excitability of D1 neurons (left) and D2 neurons (right) in _ex vivo_ slices as determined by back-propagating action potentials as measured by Ca2+-sensitive fluorescence (D1: n=12 and 15 D1 neurons, dendrites: _P_=0.90, spines: _P_=0.85; D2**:** n=16 and 10 D2 neurons, dendrites, _P_=0.27, spines, _P_=0.61; two-way ANOVA). **f,** Frequency (Hz) and amplitude (pA) of sEPSPs in D1 neurons from _ex vivo_ slices shown as box and whisker plots (Frequency: n=19 cells from 5 mice and 16 cells from 5 mice, _P_=0.23, unpaired two-tailed t-test; amplitude: n=19 cells from 5 mice and 16 cells from 5 mice, _P_=0.796, unpaired two-tailed t-test with Welch’s correction). **g,** Membrane bound DRD1 protein expression normalized to total DRD1 expression as determined by _ex vivo_ brain slice biotinylation assay shown as a dot plot (n=6 mice, _P_=0.21). **h,** Generation of _Il34fl/fl;Drd1aCre/+;Drd1eGFP-L10a_ for D1 neuron specific TRAP sequencing analysis. **i,** Volcano plot shows lack of any major gene expression changes in D1 neurons in 3 month old _Il34fl/fl;Drd1aCre/+;Drd1eGFP-L10a_ mice and littermate controls as determined by differential expression analysis (DESeq2, n=3 mice each, _P_<0.05, fold change>1.5, red: upregulated, blue: downregulated). j-k, Total striatal RNA expression analysis from control and Il34fl/fl;Drd1aCre/+ mice reveals unperturbed striatum cell-type specific gene expression pattern except the expected ~50% reduction in the expression of microglia-enriched genes. j, RPKM, normalized to controls, showing pan-medium spiny neuron (MSN), D1 neuron (D1), D2 neuron (D2), interneuron (IN), astrocyte (astro), oligodendrocyte (oligo), and microglia specific genes in Il34fl/fl;Drd1aeGFPL10a and Il34fl/fl;Drd1aCre/+;Drd1aeGFPL10a mice (n=4 mice each, P2ry12: _P_=0.003, Siglech: _P_=0.001, Cx3cr1: _P_=0.01, Csf1r: _P_=0.007, Tmem119: _P_=0.005, Fcrls: _P_=0.03, unpaired two-tailed t-test). k, RPKM, normalized to controls, showing unperturbed expression of astrocyte-specific activation markers (n=4 mice each, unpaired two-tailed t-test). l, Microglia show wild-type like expression of selected microglia sensome genes, RPKMs of selected genes have been normalized to Hexb RPKM, (n=4 mice each, unpaired two-tailed t-test). The experiments shown in h-k have been independently repeated in a second cohort (n=3 mice) with identical results. For gel source data, see Supplementary Figure 1. Box and whisker plots in b, c, e, and f are shown with arithmetic median (middle line), box shows upper and lower quartile, whiskers show min-max range. Data shown as mean± s.e.m.
Extended Data Figure 6.. Microglia regulate striatal neuron synchrony and responses to D1 agonist treatment in an ADO/A1R dependent fashion.
a, Representative tile scan of coronal brain slice showing implantation of GRIN lens and AAV9.hSyn.GCaMP6s expression in the dorsal striatum. b, Increased synchrony in the dorsal medial striatum of microglia deficient mice (n=9 mice) at baseline compared with controls (n=7 mice) (treatment: P<0.0001, distance: P<0.0001, interaction: P<0.0001; Two-way ANOVA with Sidak’s multiple comparisons test). c, Bar graphs show magnitude of Ca2+ events (ΔF/F) recorded in control (black) and microglia deficient mice (grey) at baseline (left) and in response to D1 agonist (SKF81297, 3mg/kg, right) (baseline: control, n=824 cells from 7 mice; microglia deficient, n=775 cells from 9 mice, _P_=0.87; D1 agonist: control, n=995 cells from 7 mice; microglia deficient, n=1021 cells from 9 mice; _P_=0.89, unpaired two-tailed t-test). d, e, Co-administration of A1R agonist (CPA, 0.1mg/kg) with D1 agonist (SKF81297, 3mg/kg) normalizes increased neuronal activity in microglia deficient mice. Bar graphs show wild type-like frequency (per mouse, d) and magnitude (ΔF/F, e) of Ca2+ events per neuron per minute in control (black) and microglia deficient (grey) (d, control, n= 7 mice; microglia deficient, n= 9 mice, _P_=0.82, unpaired two-tailed t-test; e, control, n=387 cells from 7 mice; microglia deficient, n=305 cells from 9 mice; _P_=0.69, unpaired two-tailed t-test). f, Spatiotemporal coding of neuronal activity (baseline shown in Fig. 3c) is disrupted by D1 agonist administration (dotted line) and largely normalized by co-administration with an A1R agonist (blue line) in control (top, n=7 mice) and microglia deficient mice (left, n=9 mice). For better visualization, the distance axis was logarithmically scaled. (Control, n=7 mice: interaction: _P_=0.0012, distance: P<0.0001, treatment: P<0.0001; Microglia deficient, n=9 mice: interaction: _P_=0.0014, distance: P<0.0001, treatment: P<0.0001; Two-way ANOVA with Sidak’s multiple comparisons test). g, Bar graphs show the frequency of Ca2+ events per neuron per minute in control (left) and microglia deficient (right) mice at baseline, in response to D1 agonist (SKF81297, 3mg/kg, i.p.) alone, or in response to D1 agonist and A1R agonist treatment (CPA, 0.1mg/kg, i.p.) treatment (Control: n=332-995 cells from 7 mice, P<0.0001; Microglia deficient: n=243-1021 cells from 9 mice, P<0.0001; One-way ANOVA with Bonferroni post hoc test). h, Confirmation of CNO-mediated neuronal activation for data shown in Fig. 3h. The neuron-specific expression of GCaMP6s and hM3Dq was achieved by injecting the indicated viruses. Virally labeled thalamocortical projection neurons were identified (mCherry expression) and calcium transients were recorded at baseline, after saline injection, and after CNO injection. i, Representative traces (left) and quantification of the area under the curve (AUC) (right) of calcium transients per mouse in virally labeled neurons pre-injection, after saline injection, and after CNO injection (n=3 mice, _P_=0.0009, One-way ANOVA with Tukey’s post hoc test). j, Microglia baseline process velocity (left) and contact with synaptic boutons (right) is not affected by either the expression of the DREADD virus (red bars) or by CNO injection (5mg/kg, black bars) alone (n=3 mice, left: _P_=0.96, right, _P_=0.25, unpaired two-tailed t-test). The experiments shown in a-g are data combined from two independent imaging cohorts of mice. Box and whisker plots in c, e, and g are shown with arithmetic median (middle line), box shows upper and lower quartile, whiskers show 1.5x interquartile range. CNO: clozapine-N-oxide; Data shown as mean± s.e.m.
Extended Data Figure 7.. Microglial expression of _Entpd1/_CD39 and Nt5e/CD73 in vitro and in vivo.
a, Dot plots show normalized, ribosome-associated mRNA levels (RPKM) for _Entpd1/_CD39 (left) and Nt5e/CD73 (right) in astrocytes, neurons, and microglia from distinct brain regions of adult mice using cell-type specific TRAP sequencing (n=2, 2, 3, 6, 4, 5, 19, and 15 mice). b, CD39 surface expression on ex vivo isolated forebrain cells of Cx3cr1CreErt2/+(Litt) mice (mice express cytosolic YFP in Cx3cr1+ microglia). Percoll-purified cells were incubated with anti-CD39-AlexaFluor700 followed by FACS analysis. The histogram shows expression levels of CD39, which is almost exclusively restricted to YFP+ microglia (red) and is not found on YFP- non-microglia cells (gray) as shown previously (data is representative of three independent experiments). c, Scheme shows ex vivo isolation procedure of CD11b+ microglia following neonatal mouse forebrain tissue dissociation and Percoll enrichment for live cells. d, e, Ex vivo CD11b+ microglia isolation procedure from neonatal pups yields highly pure microglia population. d, Microglia were positively selected for by using CD11b+ magnetic bead purification and were incubated with anti-CD39-AlexaFluor700 followed by FACS analysis to assess the purity of the population. The numbers show the percentage of live (DAPI-) cells with distinct pattern of CD39 expression levels (>98% CD39+; data is representative of two independent experiments). e, Immunofluorescent analysis of purity of CD11b+ microglia isolation. Left, cells were plated on cover slips and stained for cell-type specific protein expression using antibodies specific for IBA1 (microglia), GFAP (astrocyte), OLIG2 (oligodendrocytes) or NEUN (neurons) to identify and quantify different cells within the populations in order to assess microglia purity (n=6 GFAP/IBA1 images and 6 OLIG2/NEUN/IBA1 images). Right, representative image of cover slip containing 99% pure microglia following CD11b+ isolation procedure is shown (IBA1, green; DAPI, blue). f, left, Cell lysates of increasing numbers of CD11b+ bead-purified microglia cells have been analyzed for CD39, CD73, P2RY12, and IBA1 protein expression by Western Blot analysis as indicated, 5ng of total striatal lysate from control or Nt5e−/− (CD73-deficient) mice have been used to verify CD73 antibody specificity, H3 protein expression has been used as a loading control (k= thousand, M=million; SuperSignal ECL substrate was used to visualize CD73 expression, regular ECL was used for all other proteins) Right, Whole striatal tissue lysates of control and Nt5e−/− (CD73-deficient) striatal tissue were loaded at low (5ng) and high (30ng) concentrations and analyzed for microglia-specific protein expression (CD39, CD73, P2RY12, and IBA1) by Western Blot analysis as indicated. Whole striatal tissue lysates of control and microglia deficient mice have been used to verify antibody specificity. H3 protein expression has been used as a loading control. (SuperSignal ECL substrate was used to visualize P2RY12 expression, regular ECL was used for all other proteins). Blots are representative from two independent experiments. For gel source data, see Supplementary Figure 1. Data shown as mean± s.e.m.
Extended Data Figure 8.. Microglia suppress neuronal activation via an ATP/AMP/ADO/A1R- dependent feedback mechanism.
a, Scheme for generation of mice with microglia-specific CD39 depletion by breeding Cd39fl/fl mice to Cd39fl/fl;Cx3cr1CreErt2/+(Jung) mice followed by tamoxifen-mediated Cre induction at 4-6 weeks of age. b, Dot plots show relative expression of Entpd1, Il34, and Csf1 mRNA in the striatum of Cd39fl/fl;Cx3cr1CreErt2/+ mice and littermate controls normalized to Gapdh (n=5 and 6 mice, _Entpd1: P_=0.0012, _Il34: P_=0.38, Csf1: _P_=0.22, unpaired two-tailed t-test). c, left, Representative images of striatal sections from Cd39fl/fl and Cd39fl/fl;Cx3cr1CreErt2/+ mice stained for IBA1 (microglia, green) and DAPI (nuclei, blue) (scale bar:100μm); right, dot plots show the average number of microglia per mm2 per mouse in the striatum of Cd39fl/fl and Cd39fl/fl;Cx3cr1CreErt2/+ mice (n=4 mice, _P_=0.33, unpaired two-tailed t-test with Welch’s correction for variance). d, Microglia-specific CD39 ablation leads to increased levels of neuronal PKA activity in the striatum as measured by phosphorylation levels of GLUR1 at Ser845 in striatal protein lysate from Cd39fl/fl;Cx3cr1CreErt2/+ and littermate controls, pGLUR1 levels have been normalized to total GLUR1 in each sample, (n=8 and 6 mice, _P_=0.029, two-tailed Mann-Whitney Test). e, f, Increased seizure response in Cd39fl/fl;Cx3cr1CreErt2/+: e, Dot plot shows number of stage IV-V seizures recorded within one hour in response to D1 agonist (SKF81297, 5mg/kg) (n=11 mice each, _P_=0.0004; unpaired two-tailed t-test). f, Bar graph showing percentage of mice (left) and dot plot showing number (right) of stage IV-V seizures in response to kainic acid (15mg/kg) in Cd39fl/fl;Cx3cr1CreErt2/+ mice as compared to littermate controls (n=5 and 8 mice; left, _P_=0.17, Fisher’s exact test with Yates correction, right, _P_=0.032, unpaired two-tailed t-test). g, left, Scheme for the generation of mice with a D1 neuron-specific Adora1 depletion by breeding Adora1fl/fl mice to Drd1aCre/+ mice; right, dot plots show relative expression of Adora1 mRNA in the striatum of Adora1fl/fl;Drd1aCre/+ mice and littermate controls normalized to Gapdh (n=5 and 4 mice, _P_=0.002, unpaired two-tailed t-test). h, Co-administration of A1R agonist (CPA, 0.1mg/kg) and D1 agonist (SKF81297, 5mg/kg) does not prevent the increased seizure susceptibility in Adora1fl/fl;Drd1aCre/+ mice (n=12 and 6 mice, _P_=0.009, Fisher’s exact test with Yates correction). i, Bar graph shows percentage of microglia deficient mice with seizures in response to D1 agonist alone (SKF81297, 5mg/kg, i.p.) or co-administered with an A2AR agonist (CGS21680, 0.1mg/kg, i.p.) or an A1R agonist (CPA, 0.1m/kg, i.p.) (n=9-10 mice**,** _P_=0.005, Chi-squared test with Bonferroni post hoc adjustment). j, A1R agonist administration (CPA, 0.1mg/kg) normalizes increased PKA activity in Il34fl/fl;Drd1aCre/+ mice but does not affect PKA activity in control Il34fl/fl mice as measure by phosphorylation levels of GLUR1 at Ser845 in striatal protein lysate, pGLUR1 levels have been normalized to total GLUR1 expression in each sample (Il34fl/fl mice, n=5 mice, _P_= 0.62, Il34fl/fl;Drd1aCre/+ mice, n=5 mice, _P_=0.06, unpaired two-tailed t-test). All statistical tests are two-tailed; Data shown as mean± s.e.m.
Extended Data Figure 9.. Microglia can suppress glutamate-induced cortical neuron activation in a CD39/ADO/A1R-dependent fashion in vitro.
a-d, Experimental approaches for the assessment of adenosine-mediated regulation of cortical neuron activity in vitro. Embryonic cortical neurons were cultured on Axion microelectrode array (MEA) plates which allows for continuous electrical field recordings. a, A1Rs modulate cortical neuronal activity at baseline and in response to glutamate. On day in vitro (DIV) 14, neuronal cultures were treated with vehicle, glutamate (10μM), A1R agonist (CPA, 100nM), A1R antagonist (DCPCX, 100nM), glutamate and A1R agonist, or glutamate and A1R antagonist. Dot plot shows the percentage change in mean firing rate of neurons 1 hour after treatment compared to their baseline prior to drug treatment. (n=7 wells, P<0.0001, One-way ANOVA with Tukey’s post hoc test). b, Adenosine suppresses neuronal activity via A1R activation. On DIV14, cultures were treated with vehicle, adenosine (10μM), A1R antagonist (DCPCX, 100nM), or co-treated with adenosine and A1R antagonist. Dot plot shows percentage change in mean firing rate of neurons 1 hour after treatment compared to their baseline prior to drug treatment. (n=8 wells, P<0.0001, One-way ANOVA with Tukey’s post hoc test). c, Microglia suppress neuronal activity in response to glutamate-induced activation in an A1R-dependent manner. Microglia were isolated from neonatal pups, plated onto the neuronal culture on DIV 14, and allowed to settle for 48 hrs. Mixed cultures were treated with vehicle and/or glutamate (10μM) and/or A1R antagonist (100nM) on DIV 16. Dot plot shows percentage change in mean firing rate of neurons 1 hour after treatment compared to their baseline prior to drug treatment. (left, n=12 wells, P<0.0001, right, n=4, 6, 9, and 7 wells, _P_=0.001, One-way ANOVA with Tukey’s post hoc test). d, Microglia suppress neuronal activity in a CD39-dependent manner in response to glutamate-induced activation. Microglia were isolated from neonatal pups, plated onto the neuronal culture on DIV 14, and allowed to settle for 48 hrs. Mixed cultures were pretreated with CD39 inhibitor (ARL67156, 200μM) or vehicle (30 min) and then treated with glutamate (10μM). Dot plot shows percentage change in mean firing rate of neurons 1 hour after treatment compared to the corresponding baseline neuronal activity levels prior to their baseline prior to drug treatment. (n=12, 12, 11, and 11 wells, _P_=0.0045, One-way ANOVA with Tukey’s post hoc test). Data shown as mean ± s.e.m. and representative of 2-3 independent experiments.
Extended Data Figure 10.. Reactive microglia in different neuroinflammatory and neurodegenerative conditions show a reduction in Entpd1 and P2ry12 expression that is associated with an A1R-dependent increase in D1 neuron responses.
a-g, Changes in Entpd1 and P2ry12 gene expression are shown in: a, RNA extracted from whole striatum of 6-month old control mice and Q175 (Huntington’s disease) mice (Entpd1: _P_=0.0001; P2ry12: _P_=0.004; n=8 mice, fold change and _P_-value provided in publication). b, RNA from FACS-sorted CD11b+/F4/80+ cortical and hippocampal microglia from 8.5-month old control and 5xfAD mouse model of Alzheimer’s Disease (Entpd1: _P_=0.009; P2ry12: _P_=0.0035; n=5 mice, fold change and _P_-value provided in publication). c, RNA from FACS-sorted forebrain microglia from 10-month old control and APP/PS1 Alzheimer’s disease mouse model (n=3 mice, Entpd1: _P_=0.038; P2ry12: _P_=0.023, unpaired two-tailed t-test). d, RNA from FACS-sorted FCRLS+ phagocytic and non-phagocytic microglia isolated after stereotaxic injection of apoptotic neurons (n=4 mice, Entpd1: P<0.0001; P2ry12: P<0.0001, unpaired two-tailed t-test). e, FACS-sorted FCRLS+ microglia in 24-month old control mice or APP/PS1 Alzheimer’s disease mouse model. Plaque associated microglia were identified and sorted based on CLEC7A expression (n=6 mice, Entpd1: _P_=0.01; P2ry12: P<0.0001, One-way ANOVA with Tukey’s post hoc test). f, Massively parallel single-cell RNA-seq (MARS-seq) from isolated homeostatic microglia and disease associated microglia (DAM) in 5xfAD mice Alzheimer’s disease mouse model (Entpd1: P<0.0001; P2ry12: P<0.0001; n=8,016 cells from 3 WT and 3 AD mice, fold change and _P_-value provided in publication). g, FACS-sorted CD11b+CD45int single microglia in control and LPS-injected mice (4mg/kg) (Entpd1: P<0.0001; P2ry12: P<0.0001; n=477 microglia from saline injected mice and 770 microglia from LPS injected mice, fold change and _P_-value provided in publication). h, i, Bar graphs show increased seizure susceptibility to D1 agonist administration (SKF81297, 5mg/kg, i.p.) in LPS-injected (indicated doses, i.p.) (h) and 6-8-month old 5xfAD Alzheimer’s mice (i) that is prevented by co-administration of an A1R agonist (CPA, 0.1mg/kg, i.p.) (h, n=10-22 male mice, _P_=0.032, Chi-squared test; i, n=5-10 mice per genotype, left, _P_=0.031, Fisher’s exact test with Yates correction; right, _P_=0.49, Fisher’s exact test with Yates correction). j, Scheme illustrating the model of microglia-mediated adenosine-controlled regulation of D1 neuron responses in the healthy striatum (left) and its potential dysfunction upon microglia activation during inflammatory and/or neurodegenerative diseases (right). All statistical tests are two-tailed; Data shown as mean± s.e.m.
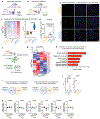
Figure 1:. Microglia respond to neuronal activation and prevent excessive neurostimulation.
a, Neuronal activity-induced transcriptional changes in microglia. CaMKII-tTa;tetO-CHRM3 mice expressing the human M3 muscarinic (hM3Dq) receptor in _CaMKII_+ neurons were bred to Cx3cr1Cre Ert2/+(Litt);Eef1a1 LSL.eGFPL10a/+ mice. Neurons were activated by clozapine-N-oxide (CNO) followed by microglia-specific mRNA analysis using translating ribosome affinity purification (TRAP). b, Gene expression changes in striatal microglia following CNO-induced activation (_z_-scored log2(RPKM); n=3 mice) c, Selected gene ontology (GO) annotations (ENRICHR) for upregulated genes (DESeq2) in striatal microglia (dotted line, _P_=0.05). d, Microglia depletion by CSF1R inhibitor PLX5622. Representative images of sagittal striatal sections from control (left) or PLX5622 treated mice (right) show nucleated (DAPI+, blue) IBA1+ (green) microglia e-g, Percentage of mice exhibiting behavioral seizures in response to i.p. injections of e, kainic acid (18mg/kg, 1hr), f, picrotoxin (1mg/kg, 30min) and g, D1 agonist (SKF81297, 5mg/kg,1hr) (e: n=9 and 10 mice, Fisher’s exact test; f: n=10 and 20 mice, Fisher’s exact test; g: n=33, 11, and 14 mice, P<0.0001, Chi-squared test with Bonferroni adjustment). The experiments shown in d, e and g have been independently repeated with same results. RPKM: reads per kilobase of transcript per million mapped reads; data shown as mean ± s.e.m
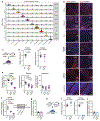
Figure 2:. Spatial control of neuronal activity by microglia.
a, Left, cell populations in the mouse striatum (t-distributed stochastic neighbour embedding (t-SNE) plot) identified by single-nuclei RNA expression analysis (15,950 nuclei). Right, Il34 (top) or Csf1 (bottom) RNA-expressing cells. b, Il34+ (top left; red) and Csf1+ cells (bottom left; red) identified by RNA in situ hybridization, DAPI+ nuclei (blue), grey matter (GM), and white matter (WM). Right, distribution of Il34+ and Csf1+ cells in GM (Il34+, 93%; Csf1+, 15%) and WM (Il34+, 7%; Csf1+, 85%) in the striatum (n = 2 and 4 mice per group, respectively). c-h, Il34fl/fl and Csf1fl/fl mice were bred to NestinCre/+ mice to generate Il34fl/fl;NestinCre/+ (purple) and Csf1fl/fl;NestinCre/+ (blue) mice. c, e, Striatal microglia numbers in GM and WM in control and mutant mice shown per frame (c: n=4 and 3 mice; unpaired two-tailed t-test e: n=4 and 2 mice) d, f, Representative images of control and mutant striatum sections showing IBA1+ (green) nucleated (DAPI+, blue) microglia. g, h, Percentages of mice with seizures in response to D1 agonist (SKF81297, 5 mg/kg) (g: n=15 and 10 mice, _P_=0.0024, Fisher’s exact test; h: n=11 and 10 mice; _P_=1.0, Fisher’s exact test). i-k, Il34fl/fl mice were bred to Drd1aCre/+ or Drd2Cre/+ mice to generate Il34fl/fl;Drd1aCre/+ (green) and Il34fl/fl;Drd2Cre/+ (grey) mice. i, Number of microglia/mm2 in cortex, striatum, and cerebellum (n=7, 4, and 3 mice, cortex: _P_=0.38; striatum: P<0.0001; cerebellum: _P_=0.14, One-way ANOVA with Tukey’s post hoc test). j-k, Percentage of mice with seizures within 1hr in response to D1 agonist (j, SKF81297, 5mg/kg) or kainic acid (k, 18mg/kg). (j: n=12, 10, and 8 mice; _P_=0.0014; k: n=17, 9, and 9 mice; _P_=0.40, Chi-squared test with Bonferroni adjustment). The experiments in g have been independently repeated in a second cohort with identical results. Data shown as mean± s.e.m.
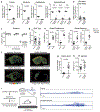
Figure 3.. Microglia control striatal neuron synchrony and firing frequency in vivo.
a, Two-photon calcium imaging of striatal neurons in control and microglia-deficient mice. b-c, Increased spatiotemporal coding of striatal neuronal activity in microglia-depleted mice at baseline. b, Representative neuron traces (n=10 neurons/mouse) and c, Correlation of spatiotemporal coding of striatal neuron activity (n=7 and 9 mice, treatment: P<0.0001, distance: P<0.0001, interaction: P<0.0001; Two-way ANOVA with Sidak’s multiple comparison test). d-e, Average frequency of Ca2+ events in control (black) and microglia-deficient (gray) mice at baseline and in response to D1 agonist (SKF81297, 3mg/kg)(n=7 and 9 mice; d, diet: _P_=0.48, treatment: _P_=0.0024, interaction: _P_=0.08, Two-way ANOVA with Tukey’s post hoc test. e, paired two-tailed t-test). f, Cumulative probability of Ca2+ events per neuron per minute in control (left) and microglia deficient (right) mice at baseline and in response to D1 agonist (3mg/kg) (n=7 and 9 mice, two-sample Kolmogorov-Smirnov test). g-h, Neuronal ATP-dependent changes in motility and localization of microglia protrusions. Using live two-photon imaging, eGFP+ microglia protrusions (Cx3cr1eGFP/+ mice) were tracked in proximity to virally-labeled tdTomato+ hM3Dq+ neurons. h, Microglial protrusion velocity per mouse (left) and mean intensity of microglial protrusions within 1μm2 of neuronal terminals per mouse (right) at baseline (saline) or upon neuronal activation (CNO, 5mg/kg, i.p.) in the absence or presence of P2RY12 antagonist (Clopidogrel, 100mg/kg, i.p.), (n= 6, 7, 6, and 3 mice, left: _P_=0.0012; right: _P_=0.01, One-way ANOVA with Tukey’s post hoc test). The data shown in c-f are data combined from two independent imaging cohorts of mice. Data shown as mean± s.e.m.
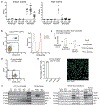
Figure 4.. Microglia suppress neuronal activation via an ATP/AMP/ADO/A1R- dependent feedback mechanism.
a, Levels of extracellular ATP, AMP, and adenosine (ADO) in primary microglia culture 60 min after addition of ATP (100μM) in the presence of CD39 inhibitor (ARL67156, 200μM) or CD73 inhibitor (APCP, 10μM) analyzed by high performance liquid chromatography (HPLC) (n=3 wells; ATP: _P_=0.0016; AMP: _P_=0.0005; ADO: _P_=0.017; One-way ANOVA with Tukey’s post hoc test). b, Left, surface expression of CD39 and CD73 in isolated forebrain cells from adult wild type (left) and CD73-deficient _Nt5e_−/− (right) mice by fluorescence-activated cell sorting (FACS). Right, expression of CD73 on CD39+ microglia (red) and CD39− non-microglia cells (blue) from wild-type mice as compared to CD39+CD73− microglia from Nt5e−/− mice (grey) (representative of three independent experiments). c, Extracellular ADO concentration in striatal dialysate from control and microglia deficient mice analyzed by mass spectrometry (n=5 mice, unpaired two-tailed t-test). d, Pharmacological and genetic dissection of the ATP-AMP-ADO-A1R circuit in microglia-dependent neuron regulation. e, PKA activity in striatal protein lysate from Il34fl/fl and Il34fl/fl;Drd1aCre/+ mice was measured by phosphorylation of GLUR1 at Ser845, DARPP32 at Thr34 (marker of D1 neuron activation), and DARPP32 at Thr75 (marker of D2 neuron activation) (Values normalized to total protein on the same blot, GLUR1Ser845: n=4 and 5 mice, two-tailed Mann-Whitney test; DARPP32Thr34: n=4, unpaired two-tailed t-test; DARPP32Thr75: n=4, unpaired two-tailed t-test). f-g, Percentage of mice showing seizures in response to D1 agonist (SKF81297, 5mg/kg) upon P2ry12 gene ablation (f, n=13 and 11 mice, Fisher’s exact test) or P2RY12 pharmacological inhibition (g, n= 16, 13, and 7 mice, _P_=0.0004, Chi-squared test with Bonferroni adjustment). h-i, Microglia-specific ablation of Entpd1/CD39. h, Striatal adenosine levels in control and Cd39fl/fl;Cx3cr1CreErt2/+ mice after D1 agonist (SKF81297, 5mg/kg) (n=5 mice, unpaired two-tailed t-test). i, Percentage of mice with seizures in response to D1 agonist alone (left) (SKF81297, 5mg/kg, n=11 mice, Fisher’s exact test) or in combination with A1R agonist (right) (CPA, 0.1mg/kg, i.p., n=5 and 9 mice, Fisher’s exact test with Yates correction). j, Percentage of mice with seizures in response to D1 agonist (SKF81297, 3mg/kg), A1R antagonist (DCPCX, 1mg/kg) or both (n=10 male mice; _P_=0.0018, Chi-squared test with Bonferroni adjustment). k, Percentage of mice with D1 neuron-specific ablation of A1R (Adora1fl/fl;Drd1aCre/+ mice) that seized in response to D1 agonist (SKF81297, 5mg/kg) (n=12 and 6 mice, Fisher’s exact test with Yates correction). l-m, Prevention of D1 agonist induced seizures by co-treatment with A1R agonist (CPA, 0.1mg/kg, i.p.) in mice following P2RY12 inhibition (l, n=10 mice, Fisher’s exact test) or striatum-specific microglia ablation (m, right, n=11 and 9 mice, D1 agonist: Fisher’s exact test; D1 agonist and A1R agonist: Fisher’s exact test). Experiments shown in f-g are combined from two independent cohorts of mice. For gel source data, see Supplementary Figure 1. All tests are two-tailed; data shown as mean± s.e.m.
Comment in
- Brain's immune cells put the brakes on neurons.
Pfeiffer T, Attwell D. Pfeiffer T, et al. Nature. 2020 Oct;586(7829):366-367. doi: 10.1038/d41586-020-02713-7. Nature. 2020. PMID: 32999439 No abstract available. - Purinergic signalling mediates the inhibitory effect of microglia on neuronal activity in the brain.
Dong YT, Tang Y. Dong YT, et al. Purinergic Signal. 2020 Dec;16(4):477-478. doi: 10.1007/s11302-020-09759-2. Epub 2021 Jan 6. Purinergic Signal. 2020. PMID: 33404957 Free PMC article. No abstract available. - Surveilling microglia dampens neuronal activity: operation of a purinergically mediated negative feedback mechanism.
Illes P, Verkhratsky A, Tang Y. Illes P, et al. Signal Transduct Target Ther. 2021 Apr 17;6(1):160. doi: 10.1038/s41392-021-00586-4. Signal Transduct Target Ther. 2021. PMID: 33866328 Free PMC article. No abstract available.
Similar articles
- The adenosine generating enzymes CD39/CD73 control microglial processes ramification in the mouse brain.
Matyash M, Zabiegalov O, Wendt S, Matyash V, Kettenmann H. Matyash M, et al. PLoS One. 2017 Apr 4;12(4):e0175012. doi: 10.1371/journal.pone.0175012. eCollection 2017. PLoS One. 2017. PMID: 28376099 Free PMC article. - Neuronal adenosine release, and not astrocytic ATP release, mediates feedback inhibition of excitatory activity.
Lovatt D, Xu Q, Liu W, Takano T, Smith NA, Schnermann J, Tieu K, Nedergaard M. Lovatt D, et al. Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6265-70. doi: 10.1073/pnas.1120997109. Epub 2012 Mar 15. Proc Natl Acad Sci U S A. 2012. PMID: 22421436 Free PMC article. - Role of the CD39/CD73 Purinergic Pathway in Modulating Arterial Thrombosis in Mice.
Covarrubias R, Chepurko E, Reynolds A, Huttinger ZM, Huttinger R, Stanfill K, Wheeler DG, Novitskaya T, Robson SC, Dwyer KM, Cowan PJ, Gumina RJ. Covarrubias R, et al. Arterioscler Thromb Vasc Biol. 2016 Sep;36(9):1809-20. doi: 10.1161/ATVBAHA.116.307374. Epub 2016 Jul 14. Arterioscler Thromb Vasc Biol. 2016. PMID: 27417582 Free PMC article. - Purinergic signaling: a potential therapeutic target for ischemic stroke.
Wang L, Li YJ, Yang X, Yang B, Zhang X, Zhang J, Zhang Q, Cheng XD, Wang JH, Yu NW. Wang L, et al. Purinergic Signal. 2023 Mar;19(1):173-183. doi: 10.1007/s11302-022-09905-y. Epub 2022 Nov 12. Purinergic Signal. 2023. PMID: 36370253 Free PMC article. Review. - The role of the CD39-CD73-adenosine pathway in liver disease.
Wang S, Gao S, Zhou D, Qian X, Luan J, Lv X. Wang S, et al. J Cell Physiol. 2021 Feb;236(2):851-862. doi: 10.1002/jcp.29932. Epub 2020 Jul 10. J Cell Physiol. 2021. PMID: 32648591 Review.
Cited by
- Monitoring and Modulating Inflammation-Associated Alterations in Synaptic Plasticity: Role of Brain Stimulation and the Blood-Brain Interface.
Lenz M, Eichler A, Vlachos A. Lenz M, et al. Biomolecules. 2021 Feb 26;11(3):359. doi: 10.3390/biom11030359. Biomolecules. 2021. PMID: 33652912 Free PMC article. Review. - Single-Cell Transcriptomics and In Situ Morphological Analyses Reveal Microglia Heterogeneity Across the Nigrostriatal Pathway.
Uriarte Huarte O, Kyriakis D, Heurtaux T, Pires-Afonso Y, Grzyb K, Halder R, Buttini M, Skupin A, Mittelbronn M, Michelucci A. Uriarte Huarte O, et al. Front Immunol. 2021 Mar 29;12:639613. doi: 10.3389/fimmu.2021.639613. eCollection 2021. Front Immunol. 2021. PMID: 33854507 Free PMC article. - Molecular and Cellular Interactions in Pathogenesis of Sporadic Parkinson Disease.
Dolgacheva LP, Zinchenko VP, Goncharov NV. Dolgacheva LP, et al. Int J Mol Sci. 2022 Oct 27;23(21):13043. doi: 10.3390/ijms232113043. Int J Mol Sci. 2022. PMID: 36361826 Free PMC article. Review. - The role of purinergic receptors in neural repair and regeneration after spinal cord injury.
Cheng RD, Ren W, Luo BY, Ye XM. Cheng RD, et al. Neural Regen Res. 2023 Aug;18(8):1684-1690. doi: 10.4103/1673-5374.363186. Neural Regen Res. 2023. PMID: 36751780 Free PMC article. Review. - Bilirubin gates the TRPM2 channel as a direct agonist to exacerbate ischemic brain damage.
Liu HW, Gong LN, Lai K, Yu XF, Liu ZQ, Li MX, Yin XL, Liang M, Shi HS, Jiang LH, Yang W, Shi HB, Wang LY, Yin SK. Liu HW, et al. Neuron. 2023 May 17;111(10):1609-1625.e6. doi: 10.1016/j.neuron.2023.02.022. Epub 2023 Mar 14. Neuron. 2023. PMID: 36921602 Free PMC article.
References
Main References
Extended References
Publication types
MeSH terms
Substances
Grants and funding
- KL2 TR002245/TR/NCATS NIH HHS/United States
- R00 DA042111/DA/NIDA NIH HHS/United States
- U01 AI066331/AI/NIAID NIH HHS/United States
- P01 HL107152/HL/NHLBI NIH HHS/United States
- R35 GM136429/GM/NIGMS NIH HHS/United States
- R01 NS106721/NS/NINDS NIH HHS/United States
- R01 NS102807/NS/NINDS NIH HHS/United States
- P01 DA047233/DA/NIDA NIH HHS/United States
- DP1 DA048931/DA/NIDA NIH HHS/United States
- RF1 MH117069/MH/NIMH NIH HHS/United States
- R01 ES025530/ES/NIEHS NIH HHS/United States
- DP2 MH100012/MH/NIMH NIH HHS/United States
- K99 NS114111/NS/NINDS NIH HHS/United States
- R01 AI126880/AI/NIAID NIH HHS/United States
- T32 AG049688/AG/NIA NIH HHS/United States
- U54 HD083211/HD/NICHD NIH HHS/United States
- R01 HL094400/HL/NHLBI NIH HHS/United States
- R21 MH115353/MH/NIMH NIH HHS/United States
- R01 GM051477/GM/NIGMS NIH HHS/United States
- R01 GM116162/GM/NIGMS NIH HHS/United States
- R01 AG045040/AG/NIA NIH HHS/United States
- R29 GM051477/GM/NIGMS NIH HHS/United States
- R01 MH118329/MH/NIMH NIH HHS/United States
- T32 AI078892/AI/NIAID NIH HHS/United States
- U01 AG058635/AG/NIA NIH HHS/United States
- R01 NS091574/NS/NINDS NIH HHS/United States
- R01 HD098363/HD/NICHD NIH HHS/United States
- K99 DA042111/DA/NIDA NIH HHS/United States
- T32 CA207021/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials