Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2 - PubMed (original) (raw)
Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2
Anamika Basu et al. Sci Rep. 2020.
Abstract
Angiotensin converting enzyme 2 (ACE2) (EC:3.4.17.23) is a transmembrane protein which is considered as a receptor for spike protein binding of novel coronavirus (SARS-CoV2). Since no specific medication is available to treat COVID-19, designing of new drug is important and essential. In this regard, in silico method plays an important role, as it is rapid and cost effective compared to the trial and error methods using experimental studies. Natural products are safe and easily available to treat coronavirus affected patients, in the present alarming situation. In this paper five phytochemicals, which belong to flavonoid and anthraquinone subclass, have been selected as small molecules in molecular docking study of spike protein of SARS-CoV2 with its human receptor ACE2 molecule. Their molecular binding sites on spike protein bound structure with its receptor have been analyzed. From this analysis, hesperidin, emodin and chrysin are selected as competent natural products from both Indian and Chinese medicinal plants, to treat COVID-19. Among them, the phytochemical hesperidin can bind with ACE2 protein and bound structure of ACE2 protein and spike protein of SARS-CoV2 noncompetitively. The binding sites of ACE2 protein for spike protein and hesperidin, are located in different parts of ACE2 protein. Ligand spike protein causes conformational change in three-dimensional structure of protein ACE2, which is confirmed by molecular docking and molecular dynamics studies. This compound modulates the binding energy of bound structure of ACE2 and spike protein. This result indicates that due to presence of hesperidin, the bound structure of ACE2 and spike protein fragment becomes unstable. As a result, this natural product can impart antiviral activity in SARS CoV2 infection. The antiviral activity of these five natural compounds are further experimentally validated with QSAR study.
Conflict of interest statement
The authors declare no competing interests.
Figures
Figure 1
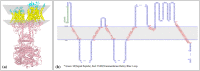
Different activities of ACE2 protein and inhibitory role of spike protein.
Figure 2
Topology of membrane protein ACE2 (a) from PDBTM (PDB ID 6M18) and (b) from MemBrain 3.1.
Figure 3
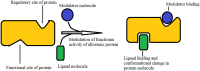
Functions of allosteric protein.
Figure 4
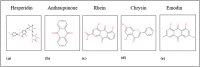
Structures of phytochemicals.
Figure 5
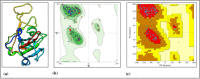
(a) 3D structure for spike protein fragment Rmachandran plot, (b) from MolProbity server, (c) PROCHECK server.
Figure 6
SARS CoV2 S protein binding with human ACE2 receptor protein (generated by using UCSF Chimera software).
Figure 7
Distorted amino acids after spike protein binding in ACE2 receptor (generated by using UCSF Chimera software).
Figure 8
Spike protein binding with ACE2 in presence of hesperidin (generated by using UCSF Chimera software).
Figure 9
Spike protein binding with ACE2 in presence of emodin (generated by using UCSF Chimera software).
Figure 10
Spike protein binding with ACE2 in presence of anthraquinone (generated by using UCSF Chimera software).
Figure 11
Rhein binding with bound spike protein fragment and ACE2 (generated by using UCSF Chimera software).
Figure 12
Chrysin binding with bound spike protein fragment and ACE2 (generated by using UCSF Chimera software).
Figure 13
Chrysin binding cleft (generated by using UCSF Chimera software).
Figure 14
Docking structure of ACE2 bound with spike protein fragment in presence of (a) chloroquine (b) hydroxychloroquine (generated by using UCSF Chimera software).
Similar articles
- A molecular docking study revealed that synthetic peptides induced conformational changes in the structure of SARS-CoV-2 spike glycoprotein, disrupting the interaction with human ACE2 receptor.
Souza PFN, Lopes FES, Amaral JL, Freitas CDT, Oliveira JTA. Souza PFN, et al. Int J Biol Macromol. 2020 Dec 1;164:66-76. doi: 10.1016/j.ijbiomac.2020.07.174. Epub 2020 Jul 18. Int J Biol Macromol. 2020. PMID: 32693122 Free PMC article. - Investigation of the genetic variation in ACE2 on the structural recognition by the novel coronavirus (SARS-CoV-2).
Guo X, Chen Z, Xia Y, Lin W, Li H. Guo X, et al. J Transl Med. 2020 Aug 24;18(1):321. doi: 10.1186/s12967-020-02486-7. J Transl Med. 2020. PMID: 32831104 Free PMC article. - In silico study of azithromycin, chloroquine and hydroxychloroquine and their potential mechanisms of action against SARS-CoV-2 infection.
Braz HLB, Silveira JAM, Marinho AD, de Moraes MEA, Moraes Filho MO, Monteiro HSA, Jorge RJB. Braz HLB, et al. Int J Antimicrob Agents. 2020 Sep;56(3):106119. doi: 10.1016/j.ijantimicag.2020.106119. Epub 2020 Jul 30. Int J Antimicrob Agents. 2020. PMID: 32738306 Free PMC article. - Angiotensin-converting enzyme 2 (ACE2) receptor and SARS-CoV-2: Potential therapeutic targeting.
Sharifkashani S, Bafrani MA, Khaboushan AS, Pirzadeh M, Kheirandish A, Yavarpour Bali H, Hessami A, Saghazadeh A, Rezaei N. Sharifkashani S, et al. Eur J Pharmacol. 2020 Oct 5;884:173455. doi: 10.1016/j.ejphar.2020.173455. Epub 2020 Jul 31. Eur J Pharmacol. 2020. PMID: 32745604 Free PMC article. Review.
Cited by
- Novel systems biology experimental pipeline reveals matairesinol's antimetastatic potential in prostate cancer: an integrated approach of network pharmacology, bioinformatics, and experimental validation.
Rajadnya R, Sharma N, Mahajan A, Ulhe A, Patil R, Hegde M, Mali A. Rajadnya R, et al. Brief Bioinform. 2024 Jul 25;25(5):bbae466. doi: 10.1093/bib/bbae466. Brief Bioinform. 2024. PMID: 39297880 Free PMC article. - Synthesis, spectroscopic, topological, hirshfeld surface analysis, and anti-covid-19 molecular docking investigation of isopropyl 1-benzoyl-4-(benzoyloxy)-2,6-diphenyl-1,2,5,6-tetrahydropyridine-3-carboxylate.
Ramalingam A, Kuppusamy M, Sambandam S, Medimagh M, Oyeneyin OE, Shanmugasundaram A, Issaoui N, Ojo ND. Ramalingam A, et al. Heliyon. 2022 Oct;8(10):e10831. doi: 10.1016/j.heliyon.2022.e10831. Epub 2022 Oct 2. Heliyon. 2022. PMID: 36211997 Free PMC article. - Exploring the Targets of Novel Corona Virus and Docking-based Screening of Potential Natural Inhibitors to Combat COVID-19.
Dey R, Samadder A, Nandi S. Dey R, et al. Curr Top Med Chem. 2022;22(29):2410-2434. doi: 10.2174/1568026623666221020163831. Curr Top Med Chem. 2022. PMID: 36281864 - Mulberry Component Kuwanon C Exerts Potent Therapeutic Efficacy In Vitro against COVID-19 by Blocking the SARS-CoV-2 Spike S1 RBD:ACE2 Receptor Interaction.
Kim YS, Kwon EB, Kim B, Chung HS, Choi G, Kim YH, Choi JG. Kim YS, et al. Int J Mol Sci. 2022 Oct 19;23(20):12516. doi: 10.3390/ijms232012516. Int J Mol Sci. 2022. PMID: 36293371 Free PMC article. - Probing the mutational landscape of the SARS-CoV-2 spike protein via quantum mechanical modeling of crystallographic structures.
Zaccaria M, Genovese L, Dawson W, Cristiglio V, Nakajima T, Johnson W, Farzan M, Momeni B. Zaccaria M, et al. PNAS Nexus. 2022 Sep 1;1(5):pgac180. doi: 10.1093/pnasnexus/pgac180. eCollection 2022 Nov. PNAS Nexus. 2022. PMID: 36712320 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous