Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria - PubMed (original) (raw)
Review
Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria
Winschau F van Zyl et al. Gut Microbes. 2020.
Abstract
Gastrointestinal (GI) diseases, and in particular those caused by bacterial infections, are a major cause of morbidity and mortality worldwide. Treatment is becoming increasingly difficult due to the increase in number of species that have developed resistance to antibiotics. Probiotic lactic acid bacteria (LAB) have considerable potential as alternatives to antibiotics, both in prophylactic and therapeutic applications. Several studies have documented a reduction, or prevention, of GI diseases by probiotic bacteria. Since the activities of probiotic bacteria are closely linked with conditions in the host's GI-tract (GIT) and changes in the population of enteric microorganisms, a deeper understanding of gut-microbial interactions is required in the selection of the most suitable probiotic. This necessitates a deeper understanding of the molecular capabilities of probiotic bacteria. In this review, we explore how probiotic microorganisms interact with enteric pathogens in the GIT. The significance of probiotic colonization and persistence in the GIT is also addressed.
Keywords: Probiotics; antimicrobial compounds; bacteriocins; colonization; competitive exclusion; enteric pathogens; gastrointestinal tract; lactic acid bacteria.
Figures
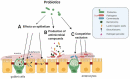
Figure 1.
Probiotic mechanisms of action against enteric pathogens in the GIT. Probiotics can affect epithelial barrier integrity by numerous mechanisms. These include: A. direct effects on the intestinal epithelial cells (IECs). Probiotics can increase the secretion of mucin glycoproteins by goblet cells that assemble into a thick mucus layer. Probiotics can augment the secretion of antimicrobial proteins (defensins) by IECs that help to eliminate commensals or pathogens that penetrate the mucus layer. Probiotics can enhance the stability of intercellular junctional complexes (tight junctions (TJ)), which decreases the intercellular permeability of IECs to pathogens and other antigens. B. Most probiotics can inhibit enteric pathogens via the production of antimicrobial substances such as bacteriocins. C. Probiotics can compete with commensals and enteric pathogens for adhesion sites in the mucus layer or IECs, thereby preventing harmful colonization and enhancing barrier function. Probiotics can alter the natural gut microbiota composition and/or gene expression, enhancing barrier integrity through the commensal microbiota. Figure created in biorender (http://biorender.io)
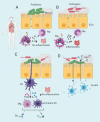
Figure 2.
Mucosal immunomodulation by probiotics in the presence of enteric pathogens. A. Down-regulation by probiotic bacteria of pro-inflammatory cytokine (IL-8) secretion in the GIT. Probiotic bacteria (or their products) may dampen an innate immune response by inhibiting the NF-ƘB inflammatory signaling pathway and influencing the production of IL-8 and subsequent recruitment of inflammatory immune cells to sites of intestinal injury. B. Activation of NF-ƘB signaling pathway by enteric pathogens, resulting in severe inflammation of intestinal epithelium tissue. C. Probiotic signaling of dendritic cells (DCs) to stimulate the secretion of anti-inflammatory cytokines such as IL-10 in response to an intestinal infection. D. Probiotics can augment the levels of IgA-secreting plasma cells in the lamina propria and promote the transcytosis of secretory IgA (sIgA) across the epithelial cell layer and secretion into the luminal mucus layer, preventing and limiting bacterial penetration of host tissues. IECs, intestinal epithelial cells; IL-8, interleukin 8; IL-10, interleukin 10; MФ, macrophage; NФ, neutrophil; NF-ƘB, nuclear factor-kappa B. TGFβ, transforming growth factor-β; Th1-3, T helper cells; Treg, regulatory T cells. Figure created in biorender (http://biorender.io)
Similar articles
- Biopreservation and probiotic potential of a large set of lactic acid bacteria isolated from Brazilian artisanal cheeses: From screening to in product approach.
Margalho LP, Jorge GP, Noleto DAP, Silva CE, Abreu JS, Piran MVF, Brocchi M, Sant'Ana AS. Margalho LP, et al. Microbiol Res. 2021 Jan;242:126622. doi: 10.1016/j.micres.2020.126622. Epub 2020 Oct 14. Microbiol Res. 2021. PMID: 33099234 - Probiotics interaction with foodborne pathogens: a potential alternative to antibiotics and future challenges.
Wan MLY, Forsythe SJ, El-Nezami H. Wan MLY, et al. Crit Rev Food Sci Nutr. 2019;59(20):3320-3333. doi: 10.1080/10408398.2018.1490885. Epub 2018 Sep 5. Crit Rev Food Sci Nutr. 2019. PMID: 29993263 Review. - Molecular genetics for probiotic engineering: dissecting lactic acid bacteria.
Suissa R, Oved R, Jankelowitz G, Turjeman S, Koren O, Kolodkin-Gal I. Suissa R, et al. Trends Microbiol. 2022 Mar;30(3):293-306. doi: 10.1016/j.tim.2021.07.007. Epub 2021 Aug 24. Trends Microbiol. 2022. PMID: 34446338 Review. - Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials.
Dunne C, Murphy L, Flynn S, O'Mahony L, O'Halloran S, Feeney M, Morrissey D, Thornton G, Fitzgerald G, Daly C, Kiely B, Quigley EM, O'Sullivan GC, Shanahan F, Collins JK. Dunne C, et al. Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):279-92. Antonie Van Leeuwenhoek. 1999. PMID: 10532384 Review. - Probiotics as drugs against human gastrointestinal infections.
Sanz Y, Nadal I, Sánchez E. Sanz Y, et al. Recent Pat Antiinfect Drug Discov. 2007 Jun;2(2):148-56. doi: 10.2174/157489107780832596. Recent Pat Antiinfect Drug Discov. 2007. PMID: 18221171 Review.
Cited by
- Applications of Probiotic-Based Multi-Components to Human, Animal and Ecosystem Health: Concepts, Methodologies, and Action Mechanisms.
Kouhounde S, Adéoti K, Mounir M, Giusti A, Refinetti P, Otu A, Effa E, Ebenso B, Adetimirin VO, Barceló JM, Thiare O, Rabetafika HN, Razafindralambo HL. Kouhounde S, et al. Microorganisms. 2022 Aug 24;10(9):1700. doi: 10.3390/microorganisms10091700. Microorganisms. 2022. PMID: 36144301 Free PMC article. Review. - Evaluation of Functional Properties of Some Lactic Acid Bacteria Strains for Probiotic Applications in Apiculture.
Urcan AC, Criste AD, Bobiș O, Cornea-Cipcigan M, Giurgiu AI, Dezmirean DS. Urcan AC, et al. Microorganisms. 2024 Jun 20;12(6):1249. doi: 10.3390/microorganisms12061249. Microorganisms. 2024. PMID: 38930631 Free PMC article. - Recent advances in genetic tools for engineering probiotic lactic acid bacteria.
Mugwanda K, Hamese S, Van Zyl WF, Prinsloo E, Du Plessis M, Dicks LMT, Thimiri Govinda Raj DB. Mugwanda K, et al. Biosci Rep. 2023 Jan 31;43(1):BSR20211299. doi: 10.1042/BSR20211299. Biosci Rep. 2023. PMID: 36597861 Free PMC article. - Biochemical, functional and genomic characterization of a new probiotic Ligilactobacillus salivarius F14 from the gut of tribes of Odisha.
Dash J, Sethi M, Deb S, Parida D, Kar S, Mahapatra S, Minz AP, Pradhan B, Prasad P, Senapati S. Dash J, et al. World J Microbiol Biotechnol. 2023 Apr 27;39(7):171. doi: 10.1007/s11274-023-03626-z. World J Microbiol Biotechnol. 2023. PMID: 37101059 - Screening of Bacteria Inhibiting Clostridium perfringens and Assessment of Their Beneficial Effects In Vitro and In Vivo with Whole Genome Sequencing Analysis.
Jiang Z, Su W, Yang M, Li W, Gong T, Zhang Y, Wen C, Wang X, Wang Y, Jin M, Lu Z. Jiang Z, et al. Microorganisms. 2022 Oct 18;10(10):2056. doi: 10.3390/microorganisms10102056. Microorganisms. 2022. PMID: 36296333 Free PMC article.
References
- Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Revs. 2012;10:66–78. - PubMed
Publication types
MeSH terms
Grants and funding
This work was supported by the National Research Foundation of South Africa (103843). WVZ received a post-doctoral fellowship from the National Research Foundation of South Africa.
LinkOut - more resources
Full Text Sources
Miscellaneous