Multi-omics-based identification of SARS-CoV-2 infection biology and candidate drugs against COVID-19 - PubMed (original) (raw)
Multi-omics-based identification of SARS-CoV-2 infection biology and candidate drugs against COVID-19
Debmalya Barh et al. Comput Biol Med. 2020 Nov.
Abstract
SARS-CoV-2 has ushered a global pandemic with no effective drug being available at present. Although several FDA-approved drugs are currently under clinical trials for drug repositioning, there is an on-going global effort for new drug identification. In this paper, using multi-omics (interactome, proteome, transcriptome, and bibliome) data and subsequent integrated analysis, we present the biological events associated with SARS-CoV-2 infection and identify several candidate drugs against this viral disease. We found that: (i) Interactome-based infection pathways differ from the other three omics-based profiles. (ii) Viral process, mRNA splicing, cytokine and interferon signaling, and ubiquitin mediated proteolysis are important pathways in SARS-CoV-2 infection. (iii) SARS-CoV-2 infection also shares pathways with Influenza A, Epstein-Barr virus, HTLV-I, Measles, and Hepatitis virus. (iv) Further, bacterial, parasitic, and protozoan infection pathways such as Tuberculosis, Malaria, and Leishmaniasis are also shared by this virus. (v) A total of 50 candidate drugs, including the prophylaxis agents and pathway specific inhibitors are identified against COVID-19. (vi) Betamethasone, Estrogen, Simvastatin, Hydrocortisone, Tositumomab, Cyclosporin A etc. are among the important drugs. (vii) Ozone, Nitric oxide, plasma components, and photosensitizer drugs are also identified as possible therapeutic candidates. (viii) Curcumin, Retinoic acids, Vitamin D, Arsenic, Copper, and Zinc may be the candidate prophylaxis agents. Nearly 70% of our identified agents are previously suggested to have anti-COVID-19 effects or under clinical trials. Among our identified drugs, the ones that are not yet tested, need validation with caution while an appropriate drug combination from these candidate drugs along with a SARS-CoV-2 specific antiviral agent is needed for effective COVID-19 management.
Keywords: COVID-19; Candidate drugs; Infection pathways; Interactome; Prophylaxis agents; Proteome; SARS-CoV-2; Transcriptome.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Authors declare no competing interest.
Figures
Graphical abstract
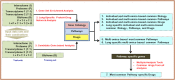
Fig. 1
The flow diagram of the overall strategy applied in this analysis. For details, see the methods section.
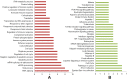
Fig. 2
Consolidated biological processes and pathways. (A) Biological processes enriched in at least 3 omics-based analyses. (B) Pathways enriched in at least 3 omics-based analyses. The X axis represents the number of appearances of the biological processes/pathways combining all of our omics-based analyses.
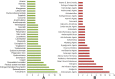
Fig. 3
Consolidated drugs and drug categories. (A) Top drugs enriched in at least 3 omics-based analyses. (B) Top drug categories with at least 2 drugs. The X axis represents the number of appearances of the drugs/drug categories combining all of our omics-based analyses.
Fig. 4
Consolidated drugs under the top ten drug categories. The X axis represents the number of appearances of the drugs under each category as per our combined all omics-based analyses.
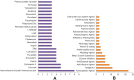
Fig. 5
Consolidated drugs and drug categories by combining the five pathways. (A) Top drugs enriched in at least 3 omics-based analyses. (B) Top drug categories with at least 2 drugs. The X axis represents the number of appearances of the drugs/drug categories combining all of our omics-based analyses.
Fig. 6
Consolidated drugs under the top nine drug categories. The X axis represents the number of appearances of the drugs under each category as per our combined all omics-based analyses.
Similar articles
- Integrative transcriptomics analysis of lung epithelial cells and identification of repurposable drug candidates for COVID-19.
Islam T, Rahman MR, Aydin B, Beklen H, Arga KY, Shahjaman M. Islam T, et al. Eur J Pharmacol. 2020 Nov 15;887:173594. doi: 10.1016/j.ejphar.2020.173594. Epub 2020 Sep 22. Eur J Pharmacol. 2020. PMID: 32971089 Free PMC article. - In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing.
Kumar Y, Singh H, Patel CN. Kumar Y, et al. J Infect Public Health. 2020 Sep;13(9):1210-1223. doi: 10.1016/j.jiph.2020.06.016. Epub 2020 Jun 16. J Infect Public Health. 2020. PMID: 32561274 Free PMC article. - Data and Text Mining Help Identify Key Proteins Involved in the Molecular Mechanisms Shared by SARS-CoV-2 and HIV-1.
Tarasova O, Ivanov S, Filimonov DA, Poroikov V. Tarasova O, et al. Molecules. 2020 Jun 26;25(12):2944. doi: 10.3390/molecules25122944. Molecules. 2020. PMID: 32604797 Free PMC article. - COVID-19: Underpinning Research for Detection, Therapeutics, and Vaccines Development.
Aljabali AAA, Bakshi HA, Satija S, Metha M, Prasher P, Ennab RM, Chellappan DK, Gupta G, Negi P, Goyal R, Sharma A, Mishra V, Dureja H, Dua K, Tambuwala MM. Aljabali AAA, et al. Pharm Nanotechnol. 2020;8(4):323-353. doi: 10.2174/2211738508999200817163335. Pharm Nanotechnol. 2020. PMID: 32811406 Review. - SARS-CoV-2-Encoded Proteome and Human Genetics: From Interaction-Based to Ribosomal Biology Impact on Disease and Risk Processes.
Sirpilla O, Bauss J, Gupta R, Underwood A, Qutob D, Freeland T, Bupp C, Carcillo J, Hartog N, Rajasekaran S, Prokop JW. Sirpilla O, et al. J Proteome Res. 2020 Nov 6;19(11):4275-4290. doi: 10.1021/acs.jproteome.0c00421. Epub 2020 Aug 10. J Proteome Res. 2020. PMID: 32686937 Review.
Cited by
- Multi-omics characterization of the monkeypox virus infection.
Huang Y, Bergant V, Grass V, Emslander Q, Hamad MS, Hubel P, Mergner J, Piras A, Krey K, Henrici A, Öllinger R, Tesfamariam YM, Dalla Rosa I, Bunse T, Sutter G, Ebert G, Schmidt FI, Way M, Rad R, Bowie AG, Protzer U, Pichlmair A. Huang Y, et al. Nat Commun. 2024 Aug 8;15(1):6778. doi: 10.1038/s41467-024-51074-6. Nat Commun. 2024. PMID: 39117661 Free PMC article. - Exploring the underlying molecular mechanisms of acute myocardial infarction after SARS-CoV-2 infection.
Xie E, Shen X, Yeo YH, Xing Z, Ebinger JE, Duan Y, Zhang Y, Cheng S, Ji F, Deng J. Xie E, et al. Am Heart J Plus. 2024 Jun 28;44:100417. doi: 10.1016/j.ahjo.2024.100417. eCollection 2024 Aug. Am Heart J Plus. 2024. PMID: 39045234 Free PMC article. - Network-based integrative multi-omics approach reveals biosignatures specific to COVID-19 disease phases.
Agamah FE, Ederveen THA, Skelton M, Martin DP, Chimusa ER, 't Hoen PAC. Agamah FE, et al. Front Mol Biosci. 2024 Jul 8;11:1393240. doi: 10.3389/fmolb.2024.1393240. eCollection 2024. Front Mol Biosci. 2024. PMID: 39040605 Free PMC article. - An RNA-Seq analysis of coronavirus in the skin of the Pangolin.
Deng S, Tian X, Belshaw R, Zhou J, Zhang S, Yang Y, Huang C, Chen W, Qiu H, Choo SW. Deng S, et al. Sci Rep. 2024 Jan 9;14(1):910. doi: 10.1038/s41598-024-51261-x. Sci Rep. 2024. PMID: 38195813 Free PMC article. - Antiviral foods in the battle against viral infections: Understanding the molecular mechanism.
Azam MS, Islam MN, Wahiduzzaman M, Alam M, Dhrubo AAK. Azam MS, et al. Food Sci Nutr. 2023 May 22;11(8):4444-4459. doi: 10.1002/fsn3.3454. eCollection 2023 Aug. Food Sci Nutr. 2023. PMID: 37576049 Free PMC article. Review.
References
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. - PMC - PubMed
- WHO . 2019. Weekly epidemiological update, coronavirus disease. (COVID-19), 28 September 2020.
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous