Canonical and Noncanonical Autophagy Pathways in Microglia - PubMed (original) (raw)
Review
. 2021 Feb 23;41(3):e0038920.
doi: 10.1128/MCB.00389-20. Epub 2020 Nov 2.
Affiliations
- PMID: 33139495
- PMCID: PMC8088277
- DOI: 10.1128/MCB.00389-20
Review
Canonical and Noncanonical Autophagy Pathways in Microglia
Julia Jülg et al. Mol Cell Biol. 2021.
Abstract
Besides the ubiquitin-proteasome system, autophagy is a major degradation pathway within cells. It delivers invading pathogens, damaged organelles, aggregated proteins, and other macromolecules from the cytosol to the lysosome for bulk degradation. This so-called canonical autophagy activity contributes to the maintenance of organelle, protein, and metabolite homeostasis as well as innate immunity. Over the past years, numerous studies rapidly deepened our knowledge on the autophagy machinery and its regulation, driven by the fact that impairment of autophagy is associated with several human pathologies, including cancer, immune diseases, and neurodegenerative disorders. Unexpectedly, components of the autophagic machinery were also found to participate in various processes that do not involve lysosomal delivery of cytosolic constituents. These functions are defined as noncanonical autophagy. Regarding neurodegenerative diseases, most research was performed in neurons, while for a long time, microglia received considerably less attention. Concomitant with the notion that microglia greatly contribute to brain health, the understanding of the role of autophagy in microglia expanded. To facilitate an overview of the current knowledge, here we present the fundamentals as well as the recent advances of canonical and noncanonical autophagy functions in microglia.
Keywords: LC3-associated endocytosis; LC3-associated phagocytosis; canonical autophagy; microglia; neurodegeneration; noncanonical autophagy.
Figures
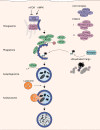
FIG 1
Autophagy pathway. The energy sensors mTORC1 and AMPK control autophagy activation via the ULK1 complex. Following activation, the ULK1 and PI3KC3 complex regulate the formation of the omegasome, and cargo is then recruited by autophagic receptors to the phagophore. Finally, the mature autophagosome fuses with lysosomes to autolysosomes, and the cargo is degraded.
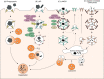
FIG 2
Overview of phagocytosis, LC3-associated phagocytosis (LAP), LC3-associated endocytosis (LANDO), and clathrin-mediated endocytosis. (A) During phagocytosis, large extracellular particles are recognized by specific receptors, engulfed by the plasma membrane, and finally internalized by phagosomes. (B) When LC3 is recruited via the PI3KC3 II complex and other autophagy proteins to the phagosome before its fusion with the lysosome, the pathway is referred to as LAP. (C) When conjugation of LC3 by autophagy proteins to Rab5- and clathrin-positive endosomes is necessary for receptor recycling, the pathway is called LANDO. (D) Clathrin-mediated endocytosis describes the uptake of smaller extracellular cargo into clathrin-coated vesicles. After uncoating, the nascent vesicle is further transported within the cell, for example, to the sorting endosome.
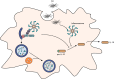
FIG 3
Selective autophagy as a possible regulator of inflammasome activity in microglia. Presence of Aβ causes activation of proinflammatory response and release of the cytokine IL-1β. Autophagy receptors p62 and NDP52 might recognize and target ubiquitinated inflammasomes for lysosomal degradation, thereby controlling Aβ-induced inflammation and survival of the cell.
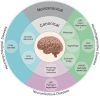
FIG 4
Canonical and noncanonical autophagy functions associated with neurological diseases. The inner and outer circles present canonical and noncanonical autophagy processes, respectively. So far, these pathways were most extensively studied in neurodegenerative diseases. However, only little is known about their functions in neuropsychological diseases. Further aspects remain to be identified. Concerning neuroinfectious diseases, either canonical xenophagy or LAP can eliminate the invading particle, depending on its pathogenic characteristics.
Similar articles
- Role of canonical and noncanonical autophagy pathways in shaping the life journey of B cells.
Wang Y, Wu L, Van Kaer L. Wang Y, et al. Front Immunol. 2024 Jul 30;15:1426204. doi: 10.3389/fimmu.2024.1426204. eCollection 2024. Front Immunol. 2024. PMID: 39139569 Free PMC article. Review. - Dying by fire: noncanonical functions of autophagy proteins in neuroinflammation and neurodegeneration.
Rickman AD, Hilyard A, Heckmann BL. Rickman AD, et al. Neural Regen Res. 2022 Feb;17(2):246-250. doi: 10.4103/1673-5374.317958. Neural Regen Res. 2022. PMID: 34269183 Free PMC article. Review. - Beyond autophagy: LC3-associated phagocytosis and endocytosis.
Peña-Martinez C, Rickman AD, Heckmann BL. Peña-Martinez C, et al. Sci Adv. 2022 Oct 28;8(43):eabn1702. doi: 10.1126/sciadv.abn1702. Epub 2022 Oct 26. Sci Adv. 2022. PMID: 36288309 Free PMC article. Review. - Autophagy Pathways in CNS Myeloid Cell Immune Functions.
Keller CW, Münz C, Lünemann JD. Keller CW, et al. Trends Neurosci. 2020 Dec;43(12):1024-1033. doi: 10.1016/j.tins.2020.09.003. Epub 2020 Sep 30. Trends Neurosci. 2020. PMID: 33010946 Review. - Human ubiquitin-like proteins as central coordinators in autophagy.
Mohan J, Wollert T. Mohan J, et al. Interface Focus. 2018 Oct 6;8(5):20180025. doi: 10.1098/rsfs.2018.0025. Epub 2018 Aug 17. Interface Focus. 2018. PMID: 30443326 Free PMC article.
Cited by
- Distinct UPR and Autophagic Functions Define Cell-Specific Responses to Proteotoxic Stress in Microglial and Neuronal Cell Lines.
Domínguez-Martín H, Gavilán E, Parrado C, Burguillos MA, Daza P, Ruano D. Domínguez-Martín H, et al. Cells. 2024 Dec 15;13(24):2069. doi: 10.3390/cells13242069. Cells. 2024. PMID: 39768160 Free PMC article. - Kaposi's sarcoma-associated herpesvirus (KSHV) utilizes the NDP52/CALCOCO2 selective autophagy receptor to disassemble processing bodies.
Robinson CA, Singh GK, Kleer M, Katsademas T, Castle EL, Boudreau BQ, Corcoran JA. Robinson CA, et al. PLoS Pathog. 2023 Jan 12;19(1):e1011080. doi: 10.1371/journal.ppat.1011080. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36634147 Free PMC article. - Expression of ALS-PFN1 impairs vesicular degradation in iPSC-derived microglia.
Funes S, Jung J, Gadd DH, Mosqueda M, Zhong J, Shankaracharya, Unger M, Stallworth K, Cameron D, Rotunno MS, Dawes P, Fowler-Magaw M, Keagle PJ, McDonough JA, Boopathy S, Sena-Esteves M, Nickerson JA, Lutz C, Skarnes WC, Lim ET, Schafer DP, Massi F, Landers JE, Bosco DA. Funes S, et al. Nat Commun. 2024 Mar 20;15(1):2497. doi: 10.1038/s41467-024-46695-w. Nat Commun. 2024. PMID: 38509062 Free PMC article. - Microglial autophagy in cerebrovascular diseases.
Chen M, Zhang H, Chu YH, Tang Y, Pang XW, Qin C, Tian DS. Chen M, et al. Front Aging Neurosci. 2022 Oct 6;14:1023679. doi: 10.3389/fnagi.2022.1023679. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36275005 Free PMC article. Review. - The role of microglial autophagy in Parkinson's disease.
Zhu R, Luo Y, Li S, Wang Z. Zhu R, et al. Front Aging Neurosci. 2022 Nov 1;14:1039780. doi: 10.3389/fnagi.2022.1039780. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36389074 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous