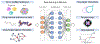Single-Cell Techniques and Deep Learning in Predicting Drug Response - PubMed (original) (raw)
Review
Single-Cell Techniques and Deep Learning in Predicting Drug Response
Zhenyu Wu et al. Trends Pharmacol Sci. 2020 Dec.
Abstract
Rapidly developing single-cell sequencing analyses produce more comprehensive profiles of the genomic, transcriptomic, and epigenomic heterogeneity of tumor subpopulations than do traditional bulk sequencing analyses. Moreover, single-cell techniques allow the response of a tumor to drug exposure to be more thoroughlyinvestigated. Deep learning (DL) models have successfully extracted features from complex bulk sequence data to predict drug responses. We review recent innovations in single-cell technologies and DL-based approaches related to drug sensitivity predictions. We believe that, by using insights from bulk sequencedata, deep transfer learning (DTL) can facilitate the use of single-cell data for training superior DL-based drug prediction models.
Keywords: deep learning models; deep transfer learning framework; drug response; single-cell technologies.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Disclaimer Statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Figure 1.. Key Figure. Combination of Single-cell and DL models in drug sensitivity prediction.
DL models are typically trained on tumor profiles, chemical and structural information of drugs and drug-target data. Extracting high-dimensional features through multi-layer perceptrons, DL models can infer drug-target interactions, purpose new drugs, and predict drug resistance.
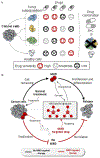
Figure 2.. Prediction of drug sensitivity at the single-cell level.
(A) Tumor subpopulations maintain diverse sensitivity to different drugs. Single usage of a drug may obtain less treatment efficiency. Knowing drug sensitivity at the single-cell level can guide the development of combination treatment that maximizes the efficiency of killing tumor cells while minimizes damage to healthy cells. (B) The MRD will proliferate and differentiate into a new tumor population which induces cancer relapse. The understanding of specific signatures characterized in MRD cells can help to discover novel drugs that specifically target MRD. MRD-targeted drug(s) administered in combination with conventional treatments can cure cancer and prevent relapse.
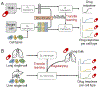
Figure 3.. Potential applications of DTL framework on single-cell data for drug sensitivity prediction.
(A) the combination usage of generative adversarial network and DTL framework transfers the drug sensitivity known at the bulk level to the single-cell level. (B) A more advanced application of DTL would transfer drug sensitivity between two single-cell data and use bulk level information as a regularizer to constrain the DL parameters.
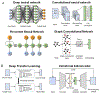
Box 1, Figure I.. The scheme of deep learning models and frameworks.
(A) Four deep learning models that have been applied in drug prediction, including DNN, CNN, RNN, and GCN. (B) Two frameworks that can be combined with DL models.
Similar articles
- Deep learning applications in single-cell genomics and transcriptomics data analysis.
Erfanian N, Heydari AA, Feriz AM, Iañez P, Derakhshani A, Ghasemigol M, Farahpour M, Razavi SM, Nasseri S, Safarpour H, Sahebkar A. Erfanian N, et al. Biomed Pharmacother. 2023 Sep;165:115077. doi: 10.1016/j.biopha.2023.115077. Epub 2023 Jul 1. Biomed Pharmacother. 2023. PMID: 37393865 Review. - Interpretation of deep learning in genomics and epigenomics.
Talukder A, Barham C, Li X, Hu H. Talukder A, et al. Brief Bioinform. 2021 May 20;22(3):bbaa177. doi: 10.1093/bib/bbaa177. Brief Bioinform. 2021. PMID: 34020542 Free PMC article. Review. - Local Epigenomic Data are more Informative than Local Genome Sequence Data in Predicting Enhancer-Promoter Interactions Using Neural Networks.
Xiao M, Zhuang Z, Pan W. Xiao M, et al. Genes (Basel). 2019 Dec 29;11(1):41. doi: 10.3390/genes11010041. Genes (Basel). 2019. PMID: 31905774 Free PMC article. - Single-Cell Genomics and Epigenomics: Technologies and Applications in Plants.
Luo C, Fernie AR, Yan J. Luo C, et al. Trends Plant Sci. 2020 Oct;25(10):1030-1040. doi: 10.1016/j.tplants.2020.04.016. Epub 2020 Jun 9. Trends Plant Sci. 2020. PMID: 32532595 Review. - Deep learning for genomics using Janggu.
Kopp W, Monti R, Tamburrini A, Ohler U, Akalin A. Kopp W, et al. Nat Commun. 2020 Jul 13;11(1):3488. doi: 10.1038/s41467-020-17155-y. Nat Commun. 2020. PMID: 32661261 Free PMC article.
Cited by
- CeDR Atlas: a knowledgebase of cellular drug response.
Wang YY, Kang H, Xu T, Hao L, Bao Y, Jia P. Wang YY, et al. Nucleic Acids Res. 2022 Jan 7;50(D1):D1164-D1171. doi: 10.1093/nar/gkab897. Nucleic Acids Res. 2022. PMID: 34634794 Free PMC article. - MMDRP: drug response prediction and biomarker discovery using multi-modal deep learning.
Taj F, Stein LD. Taj F, et al. Bioinform Adv. 2024 Jan 20;4(1):vbae010. doi: 10.1093/bioadv/vbae010. eCollection 2024. Bioinform Adv. 2024. PMID: 38371918 Free PMC article. - Deep learning methods for drug response prediction in cancer: Predominant and emerging trends.
Partin A, Brettin TS, Zhu Y, Narykov O, Clyde A, Overbeek J, Stevens RL. Partin A, et al. Front Med (Lausanne). 2023 Feb 15;10:1086097. doi: 10.3389/fmed.2023.1086097. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36873878 Free PMC article. Review. - Deep learning analysis of single-cell data in empowering clinical implementation.
Ma A, Wang J, Xu D, Ma Q. Ma A, et al. Clin Transl Med. 2022 Jul;12(7):e950. doi: 10.1002/ctm2.950. Clin Transl Med. 2022. PMID: 35858171 Free PMC article. No abstract available. - A review of computational methods for predicting cancer drug response at the single-cell level through integration with bulk RNAseq data.
Maeser D, Zhang W, Huang Y, Huang RS. Maeser D, et al. Curr Opin Struct Biol. 2024 Feb;84:102745. doi: 10.1016/j.sbi.2023.102745. Epub 2023 Dec 17. Curr Opin Struct Biol. 2024. PMID: 38109840 Review.
References
- Siegfried Z and Karni R (2018) The role of alternative splicing in cancer drug resistance. Current Opinion in Genetics & Development 48, 16–21. - PubMed
- Lee YT et al. (2018) Molecular targeted therapy: Treating cancer with specificity. European Journal of Pharmacology 834, 188–196. - PubMed
- Dagogo-Jack I and Shaw AT (2018) Tumour heterogeneity and resistance to cancer therapies. Nature Reviews Clinical Oncology 15 (2), 81–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical