UniProt: the universal protein knowledgebase in 2021 - PubMed (original) (raw)
UniProt: the universal protein knowledgebase in 2021
UniProt Consortium. Nucleic Acids Res. 2021.
Abstract
The aim of the UniProt Knowledgebase is to provide users with a comprehensive, high-quality and freely accessible set of protein sequences annotated with functional information. In this article, we describe significant updates that we have made over the last two years to the resource. The number of sequences in UniProtKB has risen to approximately 190 million, despite continued work to reduce sequence redundancy at the proteome level. We have adopted new methods of assessing proteome completeness and quality. We continue to extract detailed annotations from the literature to add to reviewed entries and supplement these in unreviewed entries with annotations provided by automated systems such as the newly implemented Association-Rule-Based Annotator (ARBA). We have developed a credit-based publication submission interface to allow the community to contribute publications and annotations to UniProt entries. We describe how UniProtKB responded to the COVID-19 pandemic through expert curation of relevant entries that were rapidly made available to the research community through a dedicated portal. UniProt resources are available under a CC-BY (4.0) license via the web at https://www.uniprot.org/.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
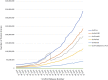
Figure 1.
Growth in the number of entries in the UniProt databases over the last decade.
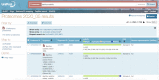
Figure 2.
Bacillus subtilis proteomes viewed on the Proteomes webpage with BUSCO and CPD scores. The left-hand panel suggests further option by which the user could filter the data, for example by only selecting reference proteomes.
Figure 3.
Catalytic activity comment and Rhea reaction visualized by the reaction graphic for the histone-lysine _N_-methyltransferase EHMT2 (UniProtKB:Q96KQ7).
Figure 4.
Information extracted from an entry describing Hepatitis C viral protein (UniProtKB:P27958) highlighting annotation added at the processed mature chain level, describing the p21 core protein. (PRO_0000037566).
Figure 5.
Community contribution. (i) Use ‘Add a publication’ functionality (red box) in the UniProtKB entry. (ii) Partial snapshot of the submission form, a sample available here:
https://community.uniprot.org/bbsub/sampleform.html
. (iii) After submission and review the publication and information are displayed in the relevant UniProtKB entry with attribution to submitter (red box) in a future public release.
Figure 6.
(A) The UniProtKB interaction viewer as seen in entry UniProtKB:Q9NSA3, the beta-catenin-interacting protein 1. (B) The interaction viewer reusable web component in the Nightingale library.
Similar articles
- UniProt: the Universal Protein Knowledgebase in 2023.
UniProt Consortium. UniProt Consortium. Nucleic Acids Res. 2023 Jan 6;51(D1):D523-D531. doi: 10.1093/nar/gkac1052. Nucleic Acids Res. 2023. PMID: 36408920 Free PMC article. - UniProt: a worldwide hub of protein knowledge.
UniProt Consortium. UniProt Consortium. Nucleic Acids Res. 2019 Jan 8;47(D1):D506-D515. doi: 10.1093/nar/gky1049. Nucleic Acids Res. 2019. PMID: 30395287 Free PMC article. - UniProt: the Universal Protein Knowledgebase in 2025.
UniProt Consortium. UniProt Consortium. Nucleic Acids Res. 2025 Jan 6;53(D1):D609-D617. doi: 10.1093/nar/gkae1010. Nucleic Acids Res. 2025. PMID: 39552041 Free PMC article. - From the research laboratory to the database: the Caenorhabditis elegans kinome in UniProtKB.
Zaru R, Magrane M, O'Donovan C; UniProt Consortium. Zaru R, et al. Biochem J. 2017 Feb 15;474(4):493-515. doi: 10.1042/BCJ20160991. Biochem J. 2017. PMID: 28159896 Free PMC article. Review. - UniProt and Mass Spectrometry-Based Proteomics-A 2-Way Working Relationship.
Bowler-Barnett EH, Fan J, Luo J, Magrane M, Martin MJ, Orchard S; UniProt Consortium. Bowler-Barnett EH, et al. Mol Cell Proteomics. 2023 Aug;22(8):100591. doi: 10.1016/j.mcpro.2023.100591. Epub 2023 Jun 8. Mol Cell Proteomics. 2023. PMID: 37301379 Free PMC article. Review.
Cited by
- Bioinformatics and network pharmacology discover the molecular mechanism of Liuwei Dihuang pills in treating cerebral palsy.
Wang L, Chen B, Xie D, Wang Y. Wang L, et al. Medicine (Baltimore). 2024 Oct 25;103(43):e40166. doi: 10.1097/MD.0000000000040166. Medicine (Baltimore). 2024. PMID: 39470545 Free PMC article. - High-quality chromosome-level genome assembly of female Artemia franciscana reveals sex chromosome and Hox gene organization.
Jo E, Cho M, Choi S, Lee SJ, Choi E, Kim J, Kim JY, Kwon S, Lee JH, Park H. Jo E, et al. Heliyon. 2024 Sep 28;10(19):e38687. doi: 10.1016/j.heliyon.2024.e38687. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39435060 Free PMC article. - Demystifying the Functional Role of Nuclear Receptors in Esophageal Cancer.
Jayaprakash S, Hegde M, Girisa S, Alqahtani MS, Abbas M, Lee EHC, Yap KC, Sethi G, Kumar AP, Kunnumakkara AB. Jayaprakash S, et al. Int J Mol Sci. 2022 Sep 19;23(18):10952. doi: 10.3390/ijms231810952. Int J Mol Sci. 2022. PMID: 36142861 Free PMC article. Review. - Genetic parameters and genome-wide association study of digital cushion thickness in Holstein cows.
Barden M, Li B, Griffiths BE, Anagnostopoulos A, Bedford C, Psifidi A, Banos G, Oikonomou G. Barden M, et al. J Dairy Sci. 2022 Oct;105(10):8237-8256. doi: 10.3168/jds.2022-22035. Epub 2022 Aug 24. J Dairy Sci. 2022. PMID: 36028347 Free PMC article. - Delineation of biomarkers and molecular pathways of residual effects of fluoxetine treatment in juvenile rhesus monkeys by proteomic profiling.
Yan Y, Park DI, Horn A, Golub M, Turck CW. Yan Y, et al. Zool Res. 2023 Jan 18;44(1):30-42. doi: 10.24272/j.issn.2095-8137.2022.196. Zool Res. 2023. PMID: 36266933 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- RG/13/5/30112/BHF_/British Heart Foundation/United Kingdom
- R01 GM080646/GM/NIGMS NIH HHS/United States
- U41 HG002273/HG/NHGRI NIH HHS/United States
- P20 GM103446/GM/NIGMS NIH HHS/United States
- U24 HG007822/HG/NHGRI NIH HHS/United States
- BB/S01781X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources