Generating high-fidelity synthetic patient data for assessing machine learning healthcare software - PubMed (original) (raw)
Generating high-fidelity synthetic patient data for assessing machine learning healthcare software
Allan Tucker et al. NPJ Digit Med. 2020.
Abstract
There is a growing demand for the uptake of modern artificial intelligence technologies within healthcare systems. Many of these technologies exploit historical patient health data to build powerful predictive models that can be used to improve diagnosis and understanding of disease. However, there are many issues concerning patient privacy that need to be accounted for in order to enable this data to be better harnessed by all sectors. One approach that could offer a method of circumventing privacy issues is the creation of realistic synthetic data sets that capture as many of the complexities of the original data set (distributions, non-linear relationships, and noise) but that does not actually include any real patient data. While previous research has explored models for generating synthetic data sets, here we explore the integration of resampling, probabilistic graphical modelling, latent variable identification, and outlier analysis for producing realistic synthetic data based on UK primary care patient data. In particular, we focus on handling missingness, complex interactions between variables, and the resulting sensitivity analysis statistics from machine learning classifiers, while quantifying the risks of patient re-identification from synthetic datapoints. We show that, through our approach of integrating outlier analysis with graphical modelling and resampling, we can achieve synthetic data sets that are not significantly different from original ground truth data in terms of feature distributions, feature dependencies, and sensitivity analysis statistics when inferring machine learning classifiers. What is more, the risk of generating synthetic data that is identical or very similar to real patients is shown to be low.
Conflict of interest statement
The authors declare no competing interests.
Figures
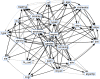
Fig. 1. Resultant graph structure for BNs learnt from samples of ground truth data.
Confidences of 100% are represented by black arcs while those <100% are represented by varying widths in grey.
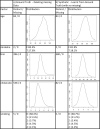
Fig. 2
Plots of sample distributions and statistics of the original ground truth data when all missing data are deleted along with plots, distributions, and statistics from the synthetic data that are generated using a BN inferred from the ground truth.
Fig. 3
Plots of sample distributions and statistics of the original ground truth data including missing data as well as plots for the synthetic data that models missing data with “Miss Nodes/States” and with latent variables.
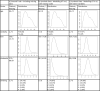
Fig. 4
Five-sample sensitivity analyses for a Bayesian generalised linear classifier on GT and SYN data (latent model) for fixed sample size of 100,000, including ROC and PR curves, and AUC and Granger statistics.
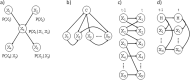
Fig. 5. Bayesian network architectures.
a A Bayesian network with four nodes. b A Bayesian network classifier with class node C. c A dynamic Bayesian network with two time-slices, t and _t_−1. d A Hidden Markov model with latent variable H.
Fig. 6. Methods to capture missing data and unmeasured effects.
a A binary “Miss Node” pointing to all continuous nodes in a Bayesian network. b A “Miss State” for discrete nodes. c A latent variable with m states to capture Missing Not at Random data and other unmeasured effects (in both discrete and continuous nodes).
Similar articles
- Towards effective machine learning in medical imaging analysis: A novel approach and expert evaluation of high-grade glioma 'ground truth' simulation on MRI.
Sepehri K, Song X, Proulx R, Hajra SG, Dobberthien B, Liu CC, D'Arcy RCN, Murray D, Krauze AV. Sepehri K, et al. Int J Med Inform. 2021 Feb;146:104348. doi: 10.1016/j.ijmedinf.2020.104348. Epub 2020 Nov 27. Int J Med Inform. 2021. PMID: 33285357 - The future of Cochrane Neonatal.
Soll RF, Ovelman C, McGuire W. Soll RF, et al. Early Hum Dev. 2020 Nov;150:105191. doi: 10.1016/j.earlhumdev.2020.105191. Epub 2020 Sep 12. Early Hum Dev. 2020. PMID: 33036834 - SynSys: A Synthetic Data Generation System for Healthcare Applications.
Dahmen J, Cook D. Dahmen J, et al. Sensors (Basel). 2019 Mar 8;19(5):1181. doi: 10.3390/s19051181. Sensors (Basel). 2019. PMID: 30857130 Free PMC article. - ARTIFICIAL INTELLIGENCE IN MEDICAL PRACTICE: REGULATIVE ISSUES AND PERSPECTIVES.
Pashkov VM, Harkusha AO, Harkusha YO. Pashkov VM, et al. Wiad Lek. 2020;73(12 cz 2):2722-2727. Wiad Lek. 2020. PMID: 33611272 Review. - A review on utilizing machine learning technology in the fields of electronic emergency triage and patient priority systems in telemedicine: Coherent taxonomy, motivations, open research challenges and recommendations for intelligent future work.
Salman OH, Taha Z, Alsabah MQ, Hussein YS, Mohammed AS, Aal-Nouman M. Salman OH, et al. Comput Methods Programs Biomed. 2021 Sep;209:106357. doi: 10.1016/j.cmpb.2021.106357. Epub 2021 Aug 16. Comput Methods Programs Biomed. 2021. PMID: 34438223 Review.
Cited by
- Comparison of Synthetic Data Generation Techniques for Control Group Survival Data in Oncology Clinical Trials: Simulation Study.
Akiya I, Ishihara T, Yamamoto K. Akiya I, et al. JMIR Med Inform. 2024 Jun 18;12:e55118. doi: 10.2196/55118. JMIR Med Inform. 2024. PMID: 38889082 Free PMC article. - Simulating complex patient populations with hierarchical learning effects to support methods development for post-market surveillance.
Davis SE, Ssemaganda H, Koola JD, Mao J, Westerman D, Speroff T, Govindarajulu US, Ramsay CR, Sedrakyan A, Ohno-Machado L, Resnic FS, Matheny ME. Davis SE, et al. BMC Med Res Methodol. 2023 Apr 11;23(1):89. doi: 10.1186/s12874-023-01913-9. BMC Med Res Methodol. 2023. PMID: 37041457 Free PMC article. - SASC: A simple approach to synthetic cohorts for generating longitudinal observational patient cohorts from COVID-19 clinical data.
Khorchani T, Gadiya Y, Witt G, Lanzillotta D, Claussen C, Zaliani A. Khorchani T, et al. Patterns (N Y). 2022 Apr 8;3(4):100453. doi: 10.1016/j.patter.2022.100453. Epub 2022 Feb 9. Patterns (N Y). 2022. PMID: 35156066 Free PMC article. - Simulation of a machine learning enabled learning health system for risk prediction using synthetic patient data.
Chen A, Chen DO. Chen A, et al. Sci Rep. 2022 Oct 26;12(1):17917. doi: 10.1038/s41598-022-23011-4. Sci Rep. 2022. PMID: 36289292 Free PMC article. - Data augmentation for generating synthetic electrogastrogram time series.
Miljković N, Milenić N, Popović NB, Sodnik J. Miljković N, et al. Med Biol Eng Comput. 2024 Sep;62(9):2879-2891. doi: 10.1007/s11517-024-03112-0. Epub 2024 May 6. Med Biol Eng Comput. 2024. PMID: 38705957 Free PMC article.
References
- The Lancet Editorial. Personalised medicine in the UK. Lancet, 391, e1 (2018). - PubMed
- FDA. Proposed Regulatory Framework for Modification to Artificial Intelligence / Machine Learning (AI/ML)–Based Software as a Medical Device (SaMD). https://www.fda.gov/media/122535/download (2020).
- Goodman, B. & Flaxman, S. European Union regulations on algorithmic decision-making and a right to explanation. Preprint at http://arxiv.org/abs/1606.08813 (2016).
- BBC 2017. Google DeepMind NHS app test broke UK privacy law. https://www.bbc.co.uk/news/technology-40483202 (2017).
- Wachter, S., Mittelstadt, B. & Floridi, L. Why a right to explanation of automated decision-making does not exist in the general data protection regulation, International Data Privacy Law. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=2903469 (2016).
LinkOut - more resources
Full Text Sources
Other Literature Sources