Staying true to yourself: mechanisms of DNA methylation maintenance in mammals - PubMed (original) (raw)
Review
Staying true to yourself: mechanisms of DNA methylation maintenance in mammals
Nataliya Petryk et al. Nucleic Acids Res. 2021.
Abstract
DNA methylation is essential to development and cellular physiology in mammals. Faulty DNA methylation is frequently observed in human diseases like cancer and neurological disorders. Molecularly, this epigenetic mark is linked to other chromatin modifications and it regulates key genomic processes, including transcription and splicing. Each round of DNA replication generates two hemi-methylated copies of the genome. These must be converted back to symmetrically methylated DNA before the next S-phase, or the mark will fade away; therefore the maintenance of DNA methylation is essential. Mechanistically, the maintenance of this epigenetic modification takes place during and after DNA replication, and occurs within the very dynamic context of chromatin re-assembly. Here, we review recent discoveries and unresolved questions regarding the mechanisms, dynamics and fidelity of DNA methylation maintenance in mammals. We also discuss how it could be regulated in normal development and misregulated in disease.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
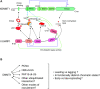
Figure 1.
The landscape and machinery of chromatin replication. (A) Replication timing along a segment of mammalian chromosome. Horizontal axis: genomic distance along the segment. Vertical axis: time at which the region is replicated during S-phase. The functionally different elements of the genome are replicated at distinct times, for instance enhancers replicate early and heterochromatin replicates late. We also indicate the typical CpG-richness of these elements (triangle at the right), and whether these CpGs are mostly unmethylated (in blue) or methylated (in red). (B) DNA methylation maintenance in the context of DNA replication and chromatin assembly. This scheme is simplified and only presents the actors mentioned in the text. Parental DNA strands are in grey, leading DNA strand in magenta, lagging DNA strand in blue. CpG dinucleotides are represented by lollipops, which are filled in white when unmethylated, and in red when methylated. The nucleosomes are shown as balls, with ‘old’ H3–H4 in orange, and new H3–H4 in white. Some of the old H3 contain H3K9me3 modifications (red flags), whereas the newly synthesized H3 do not. The DNA replication machinery generates hemimethylated CpGs.
Figure 2.
DNMT1 and UHRF1, key actors of DNA methylation maintenance. (A) Domain architecture and interactors of DNMT1 (top) and UHRF1 (bottom). Bidirectional black arrows indicate interactions. Green arrows show enzymatic modification. Lilac arrows denote inhibitory interactions. (B) Several pathways have been shown to permit DNMT1 recruitment. Additional pathways may remain to be discovered. It is not yet clear which modes of recruitment predominate in different situations.
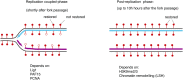
Figure 3.
Different kinetic phases of DNA methylation maintenance. The kinetics of DNA methylation maintenance combine a replication-coupled component, operating within minutes of DNA replication (left-hand panel), and a post-replication component, operating over hours (right-hand panel). The actors identified to take part in each phase are indicated at the bottom. Full red lollipops indicate the methylated 5mC and empty lollipops indicate the unmethylated (unrestored) 5mC.
Similar articles
- Acetylation of UHRF1 Regulates Hemi-methylated DNA Binding and Maintenance of Genome-wide DNA Methylation.
Hahm JY, Park JW, Kang JY, Park J, Kim CH, Kim JY, Ha NC, Kim JW, Seo SB. Hahm JY, et al. Cell Rep. 2020 Jul 28;32(4):107958. doi: 10.1016/j.celrep.2020.107958. Cell Rep. 2020. PMID: 32726623 - Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation.
Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T, Filion GJ, Deng W, de Dieuleveult M, Fritsch L, Kudithipudi S, Jeltsch A, Leonhardt H, Hajkova P, Marto JA, Arita K, Shinkai Y, Defossez PA. Ferry L, et al. Mol Cell. 2017 Aug 17;67(4):550-565.e5. doi: 10.1016/j.molcel.2017.07.012. Epub 2017 Aug 10. Mol Cell. 2017. PMID: 28803780 - Mammalian DNA methyltransferases: new discoveries and open questions.
Gowher H, Jeltsch A. Gowher H, et al. Biochem Soc Trans. 2018 Oct 19;46(5):1191-1202. doi: 10.1042/BST20170574. Epub 2018 Aug 28. Biochem Soc Trans. 2018. PMID: 30154093 Free PMC article. Review. - UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9.
Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. Liu X, et al. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. Nat Commun. 2013. PMID: 23463006 - Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.
Bronner C, Alhosin M, Hamiche A, Mousli M. Bronner C, et al. Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review.
Cited by
- Environmental exposures influence multigenerational epigenetic transmission.
Klibaner-Schiff E, Simonin EM, Akdis CA, Cheong A, Johnson MM, Karagas MR, Kirsh S, Kline O, Mazumdar M, Oken E, Sampath V, Vogler N, Wang X, Nadeau KC. Klibaner-Schiff E, et al. Clin Epigenetics. 2024 Oct 17;16(1):145. doi: 10.1186/s13148-024-01762-3. Clin Epigenetics. 2024. PMID: 39420431 Free PMC article. Review. - Replication-coupled inheritance of chromatin states.
Song A, Wang Y, Liu C, Yu J, Zhang Z, Lan L, Lin H, Zhao J, Li G. Song A, et al. Cell Insight. 2024 Aug 23;3(6):100195. doi: 10.1016/j.cellin.2024.100195. eCollection 2024 Dec. Cell Insight. 2024. PMID: 39391004 Free PMC article. Review. - The epigenetic modification of DNA methylation in neurological diseases.
Li L, Chen R, Zhang H, Li J, Huang H, Weng J, Tan H, Guo T, Wang M, Xie J. Li L, et al. Front Immunol. 2024 Sep 23;15:1401962. doi: 10.3389/fimmu.2024.1401962. eCollection 2024. Front Immunol. 2024. PMID: 39376563 Free PMC article. Review. - Epigenetic gene regulation in plants and its potential applications in crop improvement.
Zhang H, Zhu JK. Zhang H, et al. Nat Rev Mol Cell Biol. 2024 Aug 27. doi: 10.1038/s41580-024-00769-1. Online ahead of print. Nat Rev Mol Cell Biol. 2024. PMID: 39192154 Review. - Development of Locus-Directed Editing of the Epigenome from Basic Mechanistic Engineering to First Clinical Applications.
Rots MG, Jeltsch A. Rots MG, et al. Methods Mol Biol. 2024;2842:3-20. doi: 10.1007/978-1-0716-4051-7_1. Methods Mol Biol. 2024. PMID: 39012588
References
- Zemach A., McDaniel I.E., Silva P., Zilberman D.. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010; 328:916–919. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources