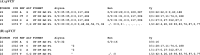Sparse Project VCF: efficient encoding of population genotype matrices - PubMed (original) (raw)
Sparse Project VCF: efficient encoding of population genotype matrices
Michael F Lin et al. Bioinformatics. 2021.
Abstract
Summary: Variant Call Format (VCF), the prevailing representation for germline genotypes in population sequencing, suffers rapid size growth as larger cohorts are sequenced and more rare variants are discovered. We present Sparse Project VCF (spVCF), an evolution of VCF with judicious entropy reduction and run-length encoding, delivering >10× size reduction for modern studies with practically minimal information loss. spVCF interoperates with VCF efficiently, including tabix-based random access. We demonstrate its effectiveness with the DiscovEHR and UK Biobank whole-exome sequencing cohorts.
Availability and implementation: Apache-licensed reference implementation: github.com/mlin/spVCF.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press.
Figures
Fig. 1.
spVCF encoding example. (A) Illustrative pVCF of four variant loci in three sequenced study participants, with matrix entries encoding called genotypes and several numeric QC measures. Some required VCF fields are omitted for brevity. (B) spVCF encoding of the same example. QC values for reference-identical and non-called cells are reduced to a power-of-two lower bound on read depth DP. Runs of identical entries down columns are abbreviated using quotation marks, then runs of these marks across rows are length-encoded. Cy’s entries are shown column-aligned for clarity; the encoded text matrix is ragged
Similar articles
- VCF-Explorer: filtering and analysing whole genome VCF files.
Akgün M, Demirci H. Akgün M, et al. Bioinformatics. 2017 Nov 1;33(21):3468-3470. doi: 10.1093/bioinformatics/btx422. Bioinformatics. 2017. PMID: 29036499 - VCF/Plotein: visualization and prioritization of genomic variants from human exome sequencing projects.
Ossio R, Garcia-Salinas OI, Anaya-Mancilla DS, Garcia-Sotelo JS, Aguilar LA, Adams DJ, Robles-Espinoza CD. Ossio R, et al. Bioinformatics. 2019 Nov 1;35(22):4803-4805. doi: 10.1093/bioinformatics/btz458. Bioinformatics. 2019. PMID: 31161195 Free PMC article. - Seqminer2: an efficient tool to query and retrieve genotypes for statistical genetics analyses from biobank scale sequence dataset.
Yang L, Jiang S, Jiang B, Liu DJ, Zhan X. Yang L, et al. Bioinformatics. 2020 Dec 8;36(19):4951-4954. doi: 10.1093/bioinformatics/btaa628. Bioinformatics. 2020. PMID: 32756942 Free PMC article. - Improved VCF normalization for accurate VCF comparison.
Bayat A, Gaëta B, Ignjatovic A, Parameswaran S. Bayat A, et al. Bioinformatics. 2017 Apr 1;33(7):964-970. doi: 10.1093/bioinformatics/btw748. Bioinformatics. 2017. PMID: 27993787 - VCF-kit: assorted utilities for the variant call format.
Cook DE, Andersen EC. Cook DE, et al. Bioinformatics. 2017 May 15;33(10):1581-1582. doi: 10.1093/bioinformatics/btx011. Bioinformatics. 2017. PMID: 28093408 Free PMC article.
Cited by
- Efficiently Summarizing Relationships in Large Samples: A General Duality Between Statistics of Genealogies and Genomes.
Ralph P, Thornton K, Kelleher J. Ralph P, et al. Genetics. 2020 Jul;215(3):779-797. doi: 10.1534/genetics.120.303253. Epub 2020 May 1. Genetics. 2020. PMID: 32357960 Free PMC article. - Lessons from national biobank projects utilizing whole-genome sequencing for population-scale genomics.
Lee H, Kim W, Kwon N, Kim C, Kim S, An JY. Lee H, et al. Genomics Inform. 2025 Mar 6;23(1):8. doi: 10.1186/s44342-025-00040-9. Genomics Inform. 2025. PMID: 40050991 Free PMC article. Review. - Analysis-ready VCF at Biobank scale using Zarr.
Czech E, Tyler W, White T, Jeffery B, Millar TR, Elsworth B, Guez J, Hancox J, Karczewski KJ, Miles A, Tallman S, Unneberg P, Wojdyla R, Zabad S, Hammerbacher J, Kelleher J. Czech E, et al. Gigascience. 2025 Jan 6;14:giaf049. doi: 10.1093/gigascience/giaf049. Gigascience. 2025. PMID: 40451243 Free PMC article. - The Scalable Variant Call Representation: Enabling Genetic Analysis Beyond One Million Genomes.
Poterba T, Vittal C, King D, Goldstein D, Goldstein JI, Schultz P, Karczewski KJ, Seed C, Neale BM. Poterba T, et al. bioRxiv [Preprint]. 2024 Jan 10:2024.01.09.574205. doi: 10.1101/2024.01.09.574205. bioRxiv. 2024. PMID: 38260295 Free PMC article. Updated. Preprint. - The scalable variant call representation: enabling genetic analysis beyond one million genomes.
Poterba T, Vittal C, King D, Goldstein D, Goldstein JI, Schultz P, Karczewski KJ, Seed C, Neale BM. Poterba T, et al. Bioinformatics. 2024 Dec 26;41(1):btae746. doi: 10.1093/bioinformatics/btae746. Bioinformatics. 2024. PMID: 39718771 Free PMC article.
References
- Danek A., Deorowicz S. (2018) GTC: how to maintain huge genotype collections in a compressed form. Bioinformatics, 34, 1834–1840. - PubMed
- Deorowicz S., Danek A. (2019) GTShark: genotype compression in large projects. Bioinformatics, 35, 4791–4793. - PubMed
- Dewey F.E. et al. (2016) Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the discovehr study. Science, 354, aaf6814. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous