Definition of a genetic relatedness cutoff to exclude recent transmission of meticillin-resistant Staphylococcus aureus: a genomic epidemiology analysis - PubMed (original) (raw)
Observational Study
Definition of a genetic relatedness cutoff to exclude recent transmission of meticillin-resistant Staphylococcus aureus: a genomic epidemiology analysis
Francesc Coll et al. Lancet Microbe. 2020 Dec.
Abstract
Background: Whole-genome sequencing (WGS) can be used in genomic epidemiology investigations to confirm or refute outbreaks of bacterial pathogens, and to support targeted and efficient infection control interventions. We aimed to define a genetic relatedness cutoff, quantified as a number of single-nucleotide polymorphisms (SNP), for meticillin-resistant Staphylococcus aureus (MRSA), above which recent (ie, within 6 months) patient-to-patient transmission could be ruled out.
Methods: We did a retrospective genomic and epidemiological analysis of MRSA data from two prospective observational cohort studies in the UK to establish SNP cutoffs for genetic relatedness, above which recent transmission was unlikely. We used three separate approaches to calculate these thresholds. First, we applied a linear mixed model to estimate the S aureus substitution rate and 95th percentile within-host diversity in a cohort in which multiple isolates were sequenced per individual. Second, we applied a simulated transmission model to this same genomic dataset. Finally, in a second cohort, we determined the genetic distance (ie, the number of SNPs) that would capture 95% of epidemiologically linked cases. We applied the three approaches to both whole-genome and core-genome sequences.
Findings: In the linear mixed model, the estimated substitution rate was roughly 5 whole-genome SNPs (wgSNPs) or 3 core-genome SNPs (cgSNPs) per genome per year, and the 95th percentile within-host diversity was 19 wgSNPs or 10 cgSNPs. The combined SNP cutoffs for detection of MRSA transmission within 6 months per this model were thus 24 wgSNPs or 13 cgSNPs. The simulated transmission model suggested that cutoffs of 17 wgSNPs or 12 cgSNPs would detect 95% of MRSA transmission events within the same timeframe. Finally, in the second cohort, cutoffs of 22 wgSNPs or 11 cgSNPs captured 95% of epidemiologically linked cases within 6 months.
Interpretation: On the basis of our results, we propose conservative cutoffs of 25 wgSNPs or 15 cgSNPS above which transmission of MRSA within the previous 6 months can be ruled out. These cutoffs could potentially be used as part of a genomic sequencing approach to the management of outbreaks of MRSA in conjunction with traditional epidemiological techniques.
Funding: UK Department of Health, Wellcome Trust, UK National Institute for Health Research.
© 2020 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license.
Figures
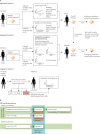
Figure 1
Overview of study design, patient cohorts, and methods (A) Approaches used to calculate a SNP cutoff above which MRSA transmission within a 6-month period is unlikely (appendix 1 pp 15–18). In approach A, a linear mixed model was used to calculate the cloud of diversity and substitution rate for MRSA; these parameters informed the estimation of the SNP cutoff. In approach B, exponential distributions of SNP distances in source and recipient patients were sampled in a simulation model (run 200 000 times) to estimate the SNP cutoff. In approach C, genomic and epidemiological data were integrated to derive the SNP cutoff. (B) Overview and relationships of all approaches and cohorts in this study. MRSA=meticillin-resistant Staphylococcus aureus. SNP=single-nucleotide polymorphism. ST=sequence type. T0=timespan 0. T1=timespan 1. Tx=generic timespan.
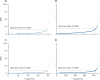
Figure 2
Empirical (A) and modelled (B) clouds of MRSA genetic diversity across individuals in the whole genome and empirical (C) and modelled (D) clouds of MRSA genetic diversity across individuals in the core genome Each datapoint corresponds to the maximum pairwise genetic distance between MRSA isolates from the same individual in cohort 1, measured as the number of SNPs in the whole or genome. The empirical cloud of diversity refers to diversity noted between isolates from the same individual collected on the same day. The modelled cloud of diversity was estimated from a linear mixed model of isolates from the same individual collected on different days. MRSA=meticillin-resistant Staphylococcus aureus. SNP=single-nucleotide polymorphism.
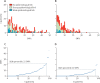
Figure 3
Distribution of SNP distances among epidemiologically linked individuals Number of individuals who had a MRSA isolate genetically linked to that of another individual in cohort 2, grouped by increasing whole-genome (A) or core-genome (B) SNP distances, and colour-coded by the strength of epidemiologically link between them (n=294). (C) and (D) show the subset of patients with strong epidemiological links (n=98). MRSA=meticillin-resistant Staphylococcus aureus. SNP=single-nucleotide polymorphism.
Similar articles
- Core genome multi-locus sequence typing as an essential tool in a high-cost livestock-associated meticillin-resistant Staphylococcus aureus CC398 hospital outbreak.
Slott Jensen ML, Nielsine Skov M, Pries Kristiansen H, Toft A, Lundgaard H, Gumpert H, Westh H, Holm A, Kolmos HJ, Kemp M. Slott Jensen ML, et al. J Hosp Infect. 2020 Apr;104(4):574-581. doi: 10.1016/j.jhin.2019.12.009. Epub 2019 Dec 16. J Hosp Infect. 2020. PMID: 31857121 - Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study.
Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. Harris SR, et al. Lancet Infect Dis. 2013 Feb;13(2):130-6. doi: 10.1016/S1473-3099(12)70268-2. Epub 2012 Nov 14. Lancet Infect Dis. 2013. PMID: 23158674 Free PMC article. - Large-Scale Evaluation of a Rapid Fully Automated Analysis Platform to Detect and Refute Outbreaks Based on MRSA Genome Comparisons.
Raven KE, Bragin E, Blane B, Leek D, Kumar N, Rhodes PA, Enoch DA, Thaxter R, Brown NM, Parkhill J, Peacock SJ. Raven KE, et al. mSphere. 2022 Dec 21;7(6):e0028322. doi: 10.1128/msphere.00283-22. Epub 2022 Oct 26. mSphere. 2022. PMID: 36286527 Free PMC article. - Contribution of whole-genome sequencing to understanding of the epidemiology and control of meticillin-resistant Staphylococcus aureus.
Humphreys H, Coleman DC. Humphreys H, et al. J Hosp Infect. 2019 Jun;102(2):189-199. doi: 10.1016/j.jhin.2019.01.025. Epub 2019 Feb 2. J Hosp Infect. 2019. PMID: 30721732 Review. - Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: Blurring of the traditional definitions.
Bal AM, Coombs GW, Holden MTG, Lindsay JA, Nimmo GR, Tattevin P, Skov RL. Bal AM, et al. J Glob Antimicrob Resist. 2016 Sep;6:95-101. doi: 10.1016/j.jgar.2016.04.004. Epub 2016 May 17. J Glob Antimicrob Resist. 2016. PMID: 27530849 Review.
Cited by
- Economic and health impact modelling of a whole genome sequencing-led intervention strategy for bacterial healthcare-associated infections for England and for the USA.
Fox JM, Saunders NJ, Jerwood SH. Fox JM, et al. Microb Genom. 2023 Aug;9(8):mgen001087. doi: 10.1099/mgen.0.001087. Microb Genom. 2023. PMID: 37555752 Free PMC article. - GraphSNP: an interactive distance viewer for investigating outbreaks and transmission networks using a graph approach.
Permana B, Beatson SA, Forde BM. Permana B, et al. BMC Bioinformatics. 2023 May 19;24(1):209. doi: 10.1186/s12859-023-05332-x. BMC Bioinformatics. 2023. PMID: 37208588 Free PMC article. - Taking hospital pathogen surveillance to the next level.
Werner G, Couto N, Feil EJ, Novais A, Hegstad K, Howden BP, Friedrich AW, Reuter S. Werner G, et al. Microb Genom. 2023 Apr;9(4):mgen001008. doi: 10.1099/mgen.0.001008. Microb Genom. 2023. PMID: 37099616 Free PMC article. - Staphylococcus aureus genotype variation among and within periprosthetic joint infections.
Ma D, Brothers KM, Maher PL, Phillips NJ, Simonetti D, William Pasculle A, Richardson AR, Cooper VS, Urish KL. Ma D, et al. J Orthop Res. 2022 Feb;40(2):420-428. doi: 10.1002/jor.25031. Epub 2021 Apr 2. J Orthop Res. 2022. PMID: 33713379 Free PMC article. - Emergence of _Cfr_-Mediated Linezolid Resistance among Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) from Healthy Pigs in Portugal.
Leão C, Clemente L, Cara d'Anjo M, Albuquerque T, Amaro A. Leão C, et al. Antibiotics (Basel). 2022 Oct 19;11(10):1439. doi: 10.3390/antibiotics11101439. Antibiotics (Basel). 2022. PMID: 36290096 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- DH_/Department of Health/United Kingdom
- 201344/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MR/S00291X/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical