Performance Characteristics of a Rapid Severe Acute Respiratory Syndrome Coronavirus 2 Antigen Detection Assay at a Public Plaza Testing Site in San Francisco - PubMed (original) (raw)
. 2021 Apr 8;223(7):1139-1144.
doi: 10.1093/infdis/jiaa802.
Paul Lebel 2, Sara Sunshine 3, Jamin Liu 3, Emily Crawford 2 4, Carina Marquez 5, Luis Rubio 5, Gabriel Chamie 5, Jackie Martinez 5, James Peng 5, Douglas Black 5, Wesley Wu 2, John Pak 2, Matthew T Laurie 3, Diane Jones 6, Steve Miller 7, Jon Jacobo 8, Susana Rojas 8, Susy Rojas 8, Robert Nakamura 9, Valerie Tulier-Laiwa 8, Maya Petersen 10, Diane V Havlir 5, Joseph DeRisi 2 3
Affiliations
- PMID: 33394052
- PMCID: PMC7799021
- DOI: 10.1093/infdis/jiaa802
Performance Characteristics of a Rapid Severe Acute Respiratory Syndrome Coronavirus 2 Antigen Detection Assay at a Public Plaza Testing Site in San Francisco
Genay Pilarowski et al. J Infect Dis. 2021.
Abstract
We evaluated the performance of the Abbott BinaxNOW rapid antigen test for coronavirus disease 2019 (Binax-CoV2) to detect virus among persons, regardless of symptoms, at a public plaza site of ongoing community transmission. Titration with cultured severe acute respiratory syndrome coronavirus 2 yielded a human observable threshold between 1.6 × 104-4.3 × 104 viral RNA copies (cycle threshold [Ct], 30.3-28.8). Among 878 subjects tested, 3% (26 of 878) were positive by reverse-transcription polymerase chain reaction, of whom 15 of 26 had a Ct <30, indicating high viral load; of these, 40% (6 of 15) were asymptomatic. Using this Ct threshold (<30) for Binax-CoV2 evaluation, the sensitivity of Binax-CoV2 was 93.3% (95% confidence interval, 68.1%-99.8%) (14 of 15) and the specificity was 99.9% (99.4%-99.9%) (855 of 856).
Keywords: COVID-19; Point of Care testing; Rapid Antigen Test; SARS-CoV-2.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures
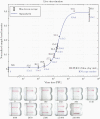
Figure 1.
Titration of in vitro grown severe acute respiratory syndrome coronavirus 2 and detection with Binax-CoV2 assay. Top, Normalized Binax-CoV2 sample band intensity (blue-green average) for cards loaded with a known amount of virus. Error bars represent standard deviation of sample band intensity of technical replicates. Reverse-transcription polymerase chain reaction (RT-PCR) testing was performed at the CLIAHUB consortium [10]. Corresponding RT-PCR cycle threshold (Ct) values (average of N and E gene probes) are shown in black, and the corresponding RNA copy numbers in blue. Note that Ct and genome copy number correlation varies by RT-PCR platform. Bottom, Representative card images from each data point. Abbreviation: PFUs, plaque-forming units.
Figure 2.
Comparison of Binax-CoV2 test with quantitative reverse-transcription polymerase chain reaction (RT-PCR) test. A, Average viral cycle threshold (Ct) values from all 26 RT-PCR–positive individuals from the community study, plotted in ascending order. Blue circles indicate Binax-CoV2–positive samples; yellow squares, Binax-CoV2–negative samples. Open symbols represent individuals who were asymptomatic on the day of the test and filled symbols, those who reported symptoms on that day. B, Normalized sample band signal from retrospective image analysis of Binax-CoV2 cards was plotted as a function of Ct value for all available scanner images (19 of 26 RT-PCR–positive samples and a random subset of RT-PCR–negative samples). Binax-CoV2 true-positives are shown in blue and labeled TP; false-negatives, shown in yellow and labeled FN; and true-negatives, shown in red and labeled TN. C, Corresponding Binax-CoV2 card images from the data in B.
Update of
- Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco.
Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, Rubio L, Chamie G, Martinez J, Peng J, Black D, Wu W, Pak J, Laurie MT, Jones D, Miller S, Jacobo J, Rojas S, Rojas S, Nakamura R, Tulier-Laiwa V, Petersen M, Havlir DV; CLIAHUB Consortium; DeRisi J. Pilarowski G, et al. medRxiv [Preprint]. 2020 Nov 12:2020.11.02.20223891. doi: 10.1101/2020.11.02.20223891. medRxiv. 2020. PMID: 33173911 Free PMC article. Updated. Preprint.
Similar articles
- Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco.
Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, Rubio L, Chamie G, Martinez J, Peng J, Black D, Wu W, Pak J, Laurie MT, Jones D, Miller S, Jacobo J, Rojas S, Rojas S, Nakamura R, Tulier-Laiwa V, Petersen M, Havlir DV; CLIAHUB Consortium; DeRisi J. Pilarowski G, et al. medRxiv [Preprint]. 2020 Nov 12:2020.11.02.20223891. doi: 10.1101/2020.11.02.20223891. medRxiv. 2020. PMID: 33173911 Free PMC article. Updated. Preprint. - Efficacy comparison of three rapid antigen tests for SARS-CoV-2 and how viral load impact their performance.
Blairon L, Cupaiolo R, Thomas I, Piteüs S, Wilmet A, Beukinga I, Tré-Hardy M. Blairon L, et al. J Med Virol. 2021 Oct;93(10):5783-5788. doi: 10.1002/jmv.27108. Epub 2021 Jun 4. J Med Virol. 2021. PMID: 34050945 Free PMC article. - Performance and Implementation Evaluation of the Abbott BinaxNOW Rapid Antigen Test in a High-Throughput Drive-Through Community Testing Site in Massachusetts.
Pollock NR, Jacobs JR, Tran K, Cranston AE, Smith S, O'Kane CY, Roady TJ, Moran A, Scarry A, Carroll M, Volinsky L, Perez G, Patel P, Gabriel S, Lennon NJ, Madoff LC, Brown C, Smole SC. Pollock NR, et al. J Clin Microbiol. 2021 Apr 20;59(5):e00083-21. doi: 10.1128/JCM.00083-21. Print 2021 Apr 20. J Clin Microbiol. 2021. PMID: 33622768 Free PMC article. - Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection.
Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, Wang S, Deng G, Zhou H, Wu Y. Diao B, et al. Clin Microbiol Infect. 2021 Feb;27(2):289.e1-289.e4. doi: 10.1016/j.cmi.2020.09.057. Epub 2020 Oct 5. Clin Microbiol Infect. 2021. PMID: 33031947 Free PMC article. - Recent Advances in Novel Lateral Flow Technologies for Detection of COVID-19.
Hsiao WW, Le TN, Pham DM, Ko HH, Chang HC, Lee CC, Sharma N, Lee CK, Chiang WH. Hsiao WW, et al. Biosensors (Basel). 2021 Aug 25;11(9):295. doi: 10.3390/bios11090295. Biosensors (Basel). 2021. PMID: 34562885 Free PMC article. Review.
Cited by
- Misdiagnosis of SARS-CoV-2: A Critical Review of the Influence of Sampling and Clinical Detection Methods.
Keaney D, Whelan S, Finn K, Lucey B. Keaney D, et al. Med Sci (Basel). 2021 May 25;9(2):36. doi: 10.3390/medsci9020036. Med Sci (Basel). 2021. PMID: 34070530 Free PMC article. Review. - Lessons From the Field: Rapid Antigen Testing Is Efficient and Practical for Mitigation of Coronavirus Disease 2019 Outbreaks in Long-Term Care Facilities.
Miller DA, Duncan L, Termini L, Prebil LA, Witt D, McCurdy SA. Miller DA, et al. Open Forum Infect Dis. 2023 Feb 21;10(2):ofad048. doi: 10.1093/ofid/ofad048. eCollection 2023 Feb. Open Forum Infect Dis. 2023. PMID: 36824624 Free PMC article. - Antigen Detection Tests for SARS-CoV-2: a systematic review and meta-analysis on real world data.
Riccò M, Ranzieri S, Peruzzi S, Valente M, Marchesi F, Bragazzi NL, Donelli D, Balzarini F, Ferraro P, Gianfredi V, Signorelli C. Riccò M, et al. Acta Biomed. 2022 May 11;93(2):e2022036. doi: 10.23750/abm.v93i2.11031. Acta Biomed. 2022. PMID: 35546034 Free PMC article. - Point-of-care COVID-19 testing in the emergency department: current status and future prospects.
May L, Tran N, Ledeboer NA. May L, et al. Expert Rev Mol Diagn. 2021 Dec;21(12):1333-1340. doi: 10.1080/14737159.2021.2005582. Epub 2021 Nov 29. Expert Rev Mol Diagn. 2021. PMID: 34758686 Free PMC article. Review. - Detection of SARS-CoV-2 variants by Abbott molecular, antigen, and serological tests.
Rodgers MA, Olivo A, Harris BJ, Lark C, Luo X, Berg MG, Meyer TV, Mohaimani A, Orf GS, Goldstein Y, Fox AS, Hirschhorn J, Glen WB, Nolte F, Landay A, Jennings C, Moy J, Servellita V, Chiu C, Batra R, Snell LB, Nebbia G, Douthwaite S, Tanuri A, Singh L, de Oliveira T, Ahouidi A, Mboup S, Cloherty GA. Rodgers MA, et al. J Clin Virol. 2022 Feb;147:105080. doi: 10.1016/j.jcv.2022.105080. Epub 2022 Jan 20. J Clin Virol. 2022. PMID: 35086043 Free PMC article.
References
- Johns Hopkins Coronavirus Resource Center. COVID-19 dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html. Accessed 23 October 2020.
- Chwe H, Quintana A, Lazer D, et al. . The state of the nation: a 50-state COVID-19 survey. Report #17: COVID-19 test result times. October 2020. http://www.kateto.net/covid19/COVID19%20CONSORTIUM%20REPORT%2017%20TESTI.... Accessed 23 October 2020.
- Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J Med 2020; 383:e120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F31 AI150007/AI/NIAID NIH HHS/United States
- T32 AI060537/AI/NIAID NIH HHS/United States
- T32 GM007810/GM/NIGMS NIH HHS/United States
- T32 AI060530/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous