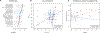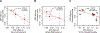Large-scale association analyses identify host factors influencing human gut microbiome composition - PubMed (original) (raw)
. 2021 Feb;53(2):156-165.
doi: 10.1038/s41588-020-00763-1. Epub 2021 Jan 18.
Carolina Medina-Gomez # 2 3, Rodrigo Bacigalupe # 4 5, Djawad Radjabzadeh # 2, Jun Wang # 4 5 6, Ayse Demirkan 7 8, Caroline I Le Roy 9, Juan Antonio Raygoza Garay 10 11, Casey T Finnicum 12, Xingrong Liu 13, Daria V Zhernakova 7 14, Marc Jan Bonder 7, Tue H Hansen 15, Fabian Frost 16, Malte C Rühlemann 17, Williams Turpin 10 11, Jee-Young Moon 18, Han-Na Kim 19 20, Kreete Lüll 21, Elad Barkan 22, Shiraz A Shah 23, Myriam Fornage 24 25, Joanna Szopinska-Tokov 26, Zachary D Wallen 27, Dmitrii Borisevich 15, Lars Agreus 28, Anna Andreasson 29, Corinna Bang 17, Larbi Bedrani 10, Jordana T Bell 9, Hans Bisgaard 23, Michael Boehnke 30, Dorret I Boomsma 31, Robert D Burk 32 33, Annique Claringbould 7, Kenneth Croitoru 10 11, Gareth E Davies 12 31, Cornelia M van Duijn 34 35, Liesbeth Duijts 3 36, Gwen Falony 4 5, Jingyuan Fu 7 37, Adriaan van der Graaf 7, Torben Hansen 15, Georg Homuth 38, David A Hughes 39 40, Richard G Ijzerman 41, Matthew A Jackson 9 42, Vincent W V Jaddoe 3 34, Marie Joossens 4 5, Torben Jørgensen 43, Daniel Keszthelyi 44 45, Rob Knight 46 47 48, Markku Laakso 49, Matthias Laudes 50, Lenore J Launer 51, Wolfgang Lieb 52, Aldons J Lusis 53 54, Ad A M Masclee 44 45, Henriette A Moll 36, Zlatan Mujagic 44 45, Qi Qibin 18, Daphna Rothschild 22, Hocheol Shin 55 56, Søren J Sørensen 57, Claire J Steves 9, Jonathan Thorsen 23, Nicholas J Timpson 39 40, Raul Y Tito 4 5, Sara Vieira-Silva 4 5, Uwe Völker 38, Henry Völzke 58, Urmo Võsa 7, Kaitlin H Wade 39 40, Susanna Walter 59 60, Kyoko Watanabe 61, Stefan Weiss 16 38, Frank U Weiss 16, Omer Weissbrod 62, Harm-Jan Westra 7, Gonneke Willemsen 31, Haydeh Payami 27, Daisy M A E Jonkers 44 45, Alejandro Arias Vasquez 26 63, Eco J C de Geus 31 64, Katie A Meyer 65 66, Jakob Stokholm 23, Eran Segal 22, Elin Org 21, Cisca Wijmenga 7, Hyung-Lae Kim 67, Robert C Kaplan 68, Tim D Spector 9, Andre G Uitterlinden 2 3 34, Fernando Rivadeneira 2 3, Andre Franke 17, Markus M Lerch 16, Lude Franke 7, Serena Sanna 7 69, Mauro D'Amato 13 70 71 72, Oluf Pedersen 15, Andrew D Paterson 73, Robert Kraaij 2, Jeroen Raes 4 5, Alexandra Zhernakova 74
Affiliations
- PMID: 33462485
- PMCID: PMC8515199
- DOI: 10.1038/s41588-020-00763-1
Large-scale association analyses identify host factors influencing human gut microbiome composition
Alexander Kurilshikov et al. Nat Genet. 2021 Feb.
Abstract
To study the effect of host genetics on gut microbiome composition, the MiBioGen consortium curated and analyzed genome-wide genotypes and 16S fecal microbiome data from 18,340 individuals (24 cohorts). Microbial composition showed high variability across cohorts: only 9 of 410 genera were detected in more than 95% of samples. A genome-wide association study of host genetic variation regarding microbial taxa identified 31 loci affecting the microbiome at a genome-wide significant (P < 5 × 10-8) threshold. One locus, the lactase (LCT) gene locus, reached study-wide significance (genome-wide association study signal: P = 1.28 × 10-20), and it showed an age-dependent association with Bifidobacterium abundance. Other associations were suggestive (1.95 × 10-10 < P < 5 × 10-8) but enriched for taxa showing high heritability and for genes expressed in the intestine and brain. A phenome-wide association study and Mendelian randomization identified enrichment of microbiome trait loci in the metabolic, nutrition and environment domains and suggested the microbiome might have causal effects in ulcerative colitis and rheumatoid arthritis.
Conflict of interest statement
Competing interests
All authors declare no competing interests.
Figures
Figure 1.. Diversity of microbiome composition across the MiBioGen cohorts.
(a) Sample size, ethnicity, genotyping array and 16S rRNA gene profiling method. The SHIP/SHIP-TREND and GEM_v12/GEM_v24/GEM_ICHIP subcohorts are combined in SHIP and GEM, respectively (Online Methods; see Supplementary Note for cohort abbreviations). This merge resulted in the total of 21 cohorts depicted in the figure. (b)* Total richness (number of genera with mean abundance over 0.1%, i.e. 10 reads out of 10,000 rarefied reads) by number of cohorts investigated. (c)* Number of core genera (genera present in >95% of samples from each cohort) by number of cohorts investigated. (d) Pearson correlation of cohort sample size with total number of genera. Confidence band represents the standard error of the regression line. (e)* Unweighted mean relative abundance of core genera across the entire MiBioGen dataset. (f)* Per-sample richness across the 21 cohorts. Asterisks indicate cohorts that differ significantly from all the others (pairwise Wilcoxon rank-sum test; FDR<0.05). (g) Diversity (Shannon index) across the 21 cohorts, with the DanFund and PNP cohorts presenting higher and lower diversity in relation to the other cohorts (pairwise Wilcoxon rank sum test; FDR<0.05). (*) For all boxplots, the central line, box and whiskers represent the median, IQR and 1.5 times the IQR.
Figure 2.. Heritability of microbiome taxa and its concordance with mbQTL mapping.
(a) Microbial taxa that showed significant heritability in the TwinsUK cohort (ACE model, nominal P<0.05, no adjustment for multiple comparison). Taxa with at least one genome-wide significant (GWS) mbQTL hit are marked red. Only taxa present in more than 10% of pairs (>17 MZ pairs, >41 DZ pairs) are shown. Circles and diamonds represent heritability value. Error bars represent 95% CI. (b) Correlation of monozygotic ICC between TwinsUK and NTR cohort. Only taxa with significant heritability (ACE model P<0.05) that are present in both TwinsUK and NTR are shown. Red and blue dots indicate bacterial taxa with/without GWS mbQTLs (P<5×10−8), respectively. Segments represent 95% CI. (c) Correlation between heritability significance (−log10PH2 TwinsUK) and the number of loci associated with microbial taxon at relaxed threshold (PmbQTL<1×10−5). Taxa with at least one GWS-associated locus are marked red. Error bars represent 95% confidence intervals.
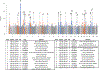
Figure 3.. Manhattan plot of the mbTL mapping meta-analysis results.
MbQTLs are indicated by letters. MbBTLs are indicated by numbers. For mbQTLs, the Spearman correlation test (two-sided) was used to identify loci that affect the covariate-adjusted abundance of bacterial taxa, excluding samples with zero abundance. For mbQTLs, p-values (two-sided) were calculated by logistic regression. Horizontal lines define nominal genome-wide significance (P=5×10−8, red) and suggestive genome-wide (P=1×10−5, blue) thresholds.
Figure 4.. Association of the LCT locus (rs182549) with the genus Bifidobacterium.
(a) Forest plot of effect sizes of rs182549 and abundance of Bifidobacterium. Effect sizes and 95% CI are defined as circles and error bars. Effect sizes were calculated from Spearman correlation p-values (Online Methods). (b) Meta-regression of the association of mean cohort age and mbQTL effect size. Confidence bands represent the standard error of the meta-regression line. (c) Meta-regression analysis of the effect of linear, squared and cubic terms of age on mbQTL effect size. Confidence bands represent the standard error of the meta-regression line. (d) Age-dependence of mbQTL effect size in the GEM cohort. Blue boxes include samples in the age range 6–16 years old. Red boxes include samples with age ≥17 years. The C/C (rs182549) genotype is a proxy of the NC_000002.11:g.136608646=(rs4988235) allele, which is associated to functional recessive hypolactasia. The central line, box and whiskers represent the median, IQR and 1.5 times the IQR, respectively. See Supplementary Note for cohort abbreviations.
Figure 5.. Phenome-wide association study (PheWAS) domain enrichment analysis.
The analysis covered top-SNPs from 30 mbTLs and 20 phenotype domains. Three thresholds for multiple testing were used: 0.05, 8.3×10−5 (Bonferroni adjustment for number of phenotypes and genotypes studied) and 5×10−8 (an arbitrary genome-wide significance threshold). Only categories with at least one significant enrichment signal are shown.
Figure 6.. Mendelian randomization (MR) analysis.
The X-axes show the SNP-exposure effect and the Y-axes show the SNP-outcome effect (SEs denoted as segments). (a) MR analysis of class Actinobacteria (exposure) and ulcerative colitis (outcome). (b) MR analysis of genus Bifidobacterium (exposure) and ulcerative colitis (outcome). (c) MR analysis of family Oxalobacteraceae (exposure) and rheumatoid arthritis (outcome).
References
- Falony G et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016). - PubMed
- Rothschild D et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24 MH068457/MH/NIMH NIH HHS/United States
- N01HC65236/HL/NHLBI NIH HHS/United States
- MR/N01183X/1/MRC_/Medical Research Council/United Kingdom
- RC2 MH089951/MH/NIMH NIH HHS/United States
- K01 HL127159/HL/NHLBI NIH HHS/United States
- N01HC65233/HL/NHLBI NIH HHS/United States
- R01 DK092127/DK/NIDDK NIH HHS/United States
- HHSN268201300005C/HL/NHLBI NIH HHS/United States
- P30 DK056350/DK/NIDDK NIH HHS/United States
- 19169/CRUK_/Cancer Research UK/United Kingdom
- U01 HG004729/HG/NHGRI NIH HHS/United States
- N01HC65226/HL/NHLBI NIH HHS/United States
- R01 MD011389/MD/NIMHD NIH HHS/United States
- N01HC65237/HL/NHLBI NIH HHS/United States
- N01HC65234/HL/NHLBI NIH HHS/United States
- RC2 MH089995/MH/NIMH NIH HHS/United States
- C18281/A19169/CRUK_/Cancer Research UK/United Kingdom
- N01HC65235/HL/NHLBI NIH HHS/United States
- R01 DK062370/DK/NIDDK NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- T32 NS095775/NS/NINDS NIH HHS/United States
- R01 HL140976/HL/NHLBI NIH HHS/United States
- MR/N030125/1/MRC_/Medical Research Council/United Kingdom
- BRC-1215-20011/DH_/Department of Health/United Kingdom
- MC_UU_00011/6/MRC_/Medical Research Council/United Kingdom
- U01 HG004446/HG/NHGRI NIH HHS/United States
- U01 DK062370/DK/NIDDK NIH HHS/United States
- UL1 TR002489/TR/NCATS NIH HHS/United States
- MC_UU_12013/3/MRC_/Medical Research Council/United Kingdom
- 204813/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 202802/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 29019/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources