Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera - PubMed (original) (raw)
. 2021 Mar 12;371(6534):1152-1153.
doi: 10.1126/science.abg6105. Epub 2021 Jan 29.
Ann-Kathrin Wallisch 1, Bianca Sänger 1, Kena A Swanson 2, Julia Mühl 1, Wei Chen 2, Hui Cai 2, Daniel Maurus 1, Ritu Sarkar 2, Özlem Türeci 1, Philip R Dormitzer 2, Uğur Şahin 3 4
Affiliations
- PMID: 33514629
- PMCID: PMC7971771
- DOI: 10.1126/science.abg6105
Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera
Alexander Muik et al. Science. 2021.
Abstract
Recently, a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage called B.1.1.7 (variant of concern: VOC 202012/01), which is reported to spread more efficiently and faster than other strains, emerged in the United Kingdom. This variant has an unusually large number of mutations, with 10 amino acid changes in the spike (S) protein, raising concerns that its recognition by neutralizing antibodies may be affected. In this study, we tested SARS-CoV-2-S pseudoviruses bearing either the Wuhan reference strain or the B.1.1.7 lineage spike protein with sera of 40 participants who were vaccinated in a previously reported trial with the messenger RNA-based COVID-19 vaccine BNT162b2. The immune sera had slightly reduced but overall largely preserved neutralizing titers against the B.1.1.7 lineage pseudovirus. These data indicate that the B.1.1.7 lineage will not escape BNT162b2-mediated protection.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
Fig. 1. 50% pseudovirus neutralization titers (pVNT50) of 40 sera from BNT162b2 vaccine recipients against VSV-SARS-CoV-2-S pseudovirus bearing the Wuhan reference strain or lineage B.1.1.7 spike protein.
Sera from n = 26 younger adults (aged 23 to 55 years; indicated by triangles) and n = 14 older adults (aged 57 to 73 years; indicated by circles) drawn at either day 29 or day 43 (7 or 21 days after vaccine dose two) were tested. Statistical significance of the difference between the neutralization of the VSV-SARS-CoV-2-S pseudovirus bearing the Wuhan or lineage B.1.1.7 spike protein was calculated by a Wilcoxon matched-pairs signed rank test. Two-tailed P values are reported. GMTs and 95% CIs are indicated.
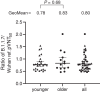
Fig. 2. pVNT50 ratio of SARS-CoV-2 lineage B.1.1.7 to Wuhan reference strain spike–pseudotyped VSV.
Triangles represent sera from younger adults (aged 23 to 55 years), and circles represent sera from older adults (aged 57 to 73 years). Sera were drawn on either day 29 or day 43 (7 or 21 days after vaccine dose two). Geometric means of the pVNT50 ratios of SARS-CoV-2 lineage B.1.1.7 to Wuhan spike–pseudotyped VSV and 95% CIs are indicated. The difference in distribution of titer ratios between younger and older adults was tested for statistical significance with a two-tailed Mann-Whitney U test.
Comment in
- Immunity to SARS-CoV-2 variants of concern.
Altmann DM, Boyton RJ, Beale R. Altmann DM, et al. Science. 2021 Mar 12;371(6534):1103-1104. doi: 10.1126/science.abg7404. Science. 2021. PMID: 33707254 No abstract available.
Similar articles
- Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera.
Muik A, Lui BG, Wallisch AK, Bacher M, Mühl J, Reinholz J, Ozhelvaci O, Beckmann N, Güimil Garcia RC, Poran A, Shpyro S, Finlayson A, Cai H, Yang Q, Swanson KA, Türeci Ö, Şahin U. Muik A, et al. Science. 2022 Feb 11;375(6581):678-680. doi: 10.1126/science.abn7591. Epub 2022 Jan 18. Science. 2022. PMID: 35040667 Free PMC article. - Serosurvey in BNT162b2 vaccine-elicited neutralizing antibodies against authentic B.1, B.1.1.7, B.1.351, B.1.525 and P.1 SARS-CoV-2 variants.
Zani A, Caccuri F, Messali S, Bonfanti C, Caruso A. Zani A, et al. Emerg Microbes Infect. 2021 Dec;10(1):1241-1243. doi: 10.1080/22221751.2021.1940305. Emerg Microbes Infect. 2021. PMID: 34092181 Free PMC article. - BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants.
Liu J, Liu Y, Xia H, Zou J, Weaver SC, Swanson KA, Cai H, Cutler M, Cooper D, Muik A, Jansen KU, Sahin U, Xie X, Dormitzer PR, Shi PY. Liu J, et al. Nature. 2021 Aug;596(7871):273-275. doi: 10.1038/s41586-021-03693-y. Epub 2021 Jun 10. Nature. 2021. PMID: 34111888 - Will Mutations in the Spike Protein of SARS-CoV-2 Lead to the Failure of COVID-19 Vaccines?
Jia Z, Gong W. Jia Z, et al. J Korean Med Sci. 2021 May 10;36(18):e124. doi: 10.3346/jkms.2021.36.e124. J Korean Med Sci. 2021. PMID: 33975397 Free PMC article. Review. - BNT162b2 mRNA COVID-19 Vaccine: First Approval.
Lamb YN. Lamb YN. Drugs. 2021 Mar;81(4):495-501. doi: 10.1007/s40265-021-01480-7. Drugs. 2021. PMID: 33683637 Free PMC article. Review.
Cited by
- Profiles of current COVID-19 vaccines.
Heinz FX, Stiasny K. Heinz FX, et al. Wien Klin Wochenschr. 2021 Apr;133(7-8):271-283. doi: 10.1007/s00508-021-01835-w. Epub 2021 Mar 16. Wien Klin Wochenschr. 2021. PMID: 33725201 Free PMC article. No abstract available. - Effects of SARS-CoV-2 variants on vaccine efficacy and response strategies.
Bian L, Gao F, Zhang J, He Q, Mao Q, Xu M, Liang Z. Bian L, et al. Expert Rev Vaccines. 2021 Apr;20(4):365-373. doi: 10.1080/14760584.2021.1903879. Epub 2021 Apr 14. Expert Rev Vaccines. 2021. PMID: 33851875 Free PMC article. Review. - The challenge of emerging SARS-CoV-2 mutants to vaccine development.
Li R, Liu J, Zhang H. Li R, et al. J Genet Genomics. 2021 Feb 20;48(2):102-106. doi: 10.1016/j.jgg.2021.03.001. Epub 2021 Apr 20. J Genet Genomics. 2021. PMID: 33994322 Free PMC article. Review. No abstract available. - Phylogenomic Evidence of Reinfection and Persistence of SARS-CoV-2: First Report from Colombia.
Ramírez JD, Muñoz M, Ballesteros N, Patiño LH, Castañeda S, Rincón CA, Mendez C, Oliveros C, Perez J, Márquez EK, Ortiz FLS, Correa-Cárdenas CA, Duque MC, Paniz-Mondolfi A. Ramírez JD, et al. Vaccines (Basel). 2021 Mar 19;9(3):282. doi: 10.3390/vaccines9030282. Vaccines (Basel). 2021. PMID: 33808687 Free PMC article. - A Comprehensive Review on the Current Vaccines and Their Efficacies to Combat SARS-CoV-2 Variants.
Rabaan AA, Mutair AA, Hajissa K, Alfaraj AH, Al-Jishi JM, Alhajri M, Alwarthan S, Alsuliman SA, Al-Najjar AH, Al Zaydani IA, Al-Absi GH, Alshaikh SA, Alkathlan MS, Almuthree SA, Alawfi A, Alshengeti A, Almubarak FZ, Qashgari MS, Abdalla ANK, Alhumaid S. Rabaan AA, et al. Vaccines (Basel). 2022 Oct 2;10(10):1655. doi: 10.3390/vaccines10101655. Vaccines (Basel). 2022. PMID: 36298520 Free PMC article. Review.
References
- Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W. Jr.., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C.; C4591001 Clinical Trial Group , Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). 10.1056/NEJMoa2034577 - DOI - PMC - PubMed
- A. Rambaut, N. Loman, O. Pybus, W. Barclay, J. Barrett, A. Carabelli, T. Connor, T. Peacock, D. L. Robertson, E. Volz; COVID-19 Genomics Consortium UK (CoG-UK), Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.org (2020); https://virological.org/t/preliminary-genomic-characterisation-of-an-eme....
- E. Volz, S. Mishra, M. Chand, J. C. Barrett, R. Johnson, L. Geidelberg, W. R. Hinsley, D. J. Laydon, G. Dabrera, Á. O’Toole, R. Amato, M. Ragonnet-Cronin, I. Harrison, B. Jackson, C. V. Ariani, O. Boyd, N. J. Loman, J. T. McCrone, S. Gonçalves, D. Jorgensen, R. Myers, V. Hill, D. K. Jackson, K. Gaythorpe, N. Groves, J. Sillitoe, D. P. Kwiatkowski, The COVID-19 Genomics UK (COG-UK) consortium, S. Flaxman, O. Ratmann, S. Bhatt, S. Hopkins, A. Gandy, A. Rambaut, N. M. Ferguson, Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv 2020.12.30.20249034 [Preprint]. 4 January 2021. 10.1101/2020.12.30.20249034.10.1101/2020.12.30.20249034 - DOI - DOI
- Starr T. N., Greaney A. J., Hilton S. K., Ellis D., Crawford K. H. D., Dingens A. S., Navarro M. J., Bowen J. E., Tortorici M. A., Walls A. C., King N. P., Veesler D., Bloom J. D., Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310.e20 (2020). 10.1016/j.cell.2020.08.012 - DOI - PMC - PubMed
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y. Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X. F., Li J., Zhang N. N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D. Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C. F., Zhou Y., Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369, 1603–1607 (2020). 10.1126/science.abc4730 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous