Brassica carinata genome characterization clarifies U's triangle model of evolution and polyploidy in Brassica - PubMed (original) (raw)
. 2021 May 27;186(1):388-406.
doi: 10.1093/plphys/kiab048.
Xiaoming Song 1 2 3, Dong Xiao 4, Ke Gong 1, Pengchuan Sun 1, Yiming Ren 4, Jiaqing Yuan 1, Tong Wu 1, Qihang Yang 1, Xinyu Li 1, Fulei Nie 1, Nan Li 1, Shuyan Feng 1, Qiaoying Pei 1, Tong Yu 1, Changwei Zhang 4, Tongkun Liu 4, Xiyin Wang 1, Jinghua Yang 5
Affiliations
- PMID: 33599732
- PMCID: PMC8154070
- DOI: 10.1093/plphys/kiab048
Brassica carinata genome characterization clarifies U's triangle model of evolution and polyploidy in Brassica
Xiaoming Song et al. Plant Physiol. 2021.
Abstract
Ethiopian mustard (Brassica carinata) in the Brassicaceae family possesses many excellent agronomic traits. Here, the high-quality genome sequence of B. carinata is reported. Characterization revealed a genome anchored to 17 chromosomes with a total length of 1.087 Gb and an N50 scaffold length of 60 Mb. Repetitive sequences account for approximately 634 Mb or 58.34% of the B. carinata genome. Notably, 51.91% of 97,149 genes are confined to the terminal 20% of chromosomes as a result of the expansion of repeats in pericentromeric regions. Brassica carinata shares one whole-genome triplication event with the five other species in U's triangle, a classic model of evolution and polyploidy in Brassica. Brassica carinata was deduced to have formed ∼0.047 Mya, which is slightly earlier than B. napus but later than B. juncea. Our analysis indicated that the relationship between the two subgenomes (BcaB and BcaC) is greater than that between other two tetraploid subgenomes (BjuB and BnaC) and their respective diploid parents. RNA-seq datasets and comparative genomic analysis were used to identify several key genes in pathways regulating disease resistance and glucosinolate metabolism. Further analyses revealed that genome triplication and tandem duplication played important roles in the expansion of those genes in Brassica species. With the genome sequencing of B. carinata completed, the genomes of all six Brassica species in U's triangle are now resolved. The data obtained from genome sequencing, transcriptome analysis, and comparative genomic efforts in this study provide valuable insights into the genome evolution of the six Brassica species in U's triangle.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
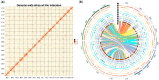
Figure 1
Hi-C map, chromosomal features, and functional annotation of the Brassica carinata genome. A, Hi-C map showing genome-wide all-by-all interactions between 17 chromosomes (B01–B08, C01–C09). B, I. 17 chromosomes of B. carinata. B01–B08 were from the BcaB subgenome, and C01–C09 were from the BcaC subgenome. II. Gene density. III–VII. Repeat sequences distribution on each chromosome: III. Gypsy; IV. Copia; V. Long interspersed nuclear elements (LINEs); VI. DNA repeat; VII. Simple sequence repeats (SSR). VIII. The gene expression was normalized as Fragments Per Kilobase of transcript sequence per Million base pairs. Gene expression levels (Log2FPKM) under drought treatment. IX. Gene expression levels (Log2FPKM) of control. X. Tandem genes distribution on each chromosome. XI. Orthologous genes distribution on each chromosome. XII. Lines connecting colinear blocks between BcaB and BcaC subgenomes, and the colors assigned according to each BcaB chromosome.
Figure 2
Divergence time estimation and recurrent genome duplications in B. carinata. A, Divergence time estimation among the six species in U’s triangle model of Brassica and Arabidopsis. The numbers on the nodes represent the Ks values and divergence time of the species (million years ago, Mya). The 95% confidence intervals of divergence time are in parentheses at each node. WGT indicates whole-genome triplication. B, Ks density plot of colinear genes for the two subgenomes (BcaB and BcaC) of B. carinata, B. nigra (Bni), B. oleracea (Bol), and Arabidopsis (Ath). C, Recurrent genome duplications in B. carinata. Genomic alignments are shown between the basal angiosperm Amborella trichopoda, the basal eudicot Vitis vinifera, the model Brassicaceae A. thaliana, and B. oleracea, B. nigra, and B. carinata.
Figure 3
Genome colinear analyses of Brassica carinata and five other Brassica species in U’s triangle. A, Global colinearity of the genomes of the six species in U’s triangle, including three diploid species, B. rapa (A genome), B. nigra (B genome), and B. oleracea (C genome) and three tetraploid species, B. napus (AACC, BnaA, and BnaC subgenomes); B. juncea (AABB, BjuA and BjuB subgenomes); and B. carinata (BBCC, BcaB and BcaC subgenomes). B, Boxplots of the relationships between two B subgenomes (BcaB and BjuB) and their corresponding diploid genomes (BniB) by calculating the Ks of colinear genes. A significance test was conducted using the R program. C, Boxplots of the relationships between two C subgenomes (BnaC and BcaC) and their corresponding diploid genomes (BolC) by calculating the Ks of colinear genes.
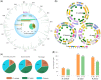
Figure 4
Tandem genes and gene conversion analyses in the two subgenomes of B. carinata. A, Chromosomal distribution of tandem genes with different Ks values in the two subgenomes of B. carinata. The diagram shows tandem gene pairs with Ka/Ks >1. The green line represents Ks <0.2, the orange line 0.2 <Ks <0.45, and the gray line Ks >0.45. B, Distribution of gene conversions between the two subgenomes of B. carinata, B. juncea, and B. napus. The connected lines of different colors between the two circles of each species indicate a converted gene. The red lines indicate the inner circle subgenome is the donor; the blue lines indicate the outer circle subgenome is the donor; the black lines indicate the gene conversion was bidirectional between two subgenomes; and the gray lines indicate the direction of the gene conversion is unknown. C, Statistics of converted gene pairs, quartet gene number, and the percentage of converted genes pairs accounting for quartet number in the three tetraploid species B. carinata, B. napus, and B. juncea. D, The bar chart showed the number of converted gene pairs between two subgenomes in three tetraploid species.
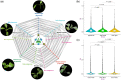
Figure 5
Colinear homologous gene expression dominance analyses between the two subgenomes of B. carinata. A, Homoeolog expression dominance analyses using the RNA-seq datasets of six tissues (top: root [left], stem, leaf [right]; bottom: flower [left], silique, seed [right]) of B. carinata. Blue and red represent the number of dominant genes in the BcaB and BcaC subgenomes, respectively; gray represents the neutral genes in the two subgenomes. B, Venn diagrams of the tissue-specific and common dominant genes in the two subgenomes (left, BcaB; right, BcaC). The pie diagrams show the enriched GO terms and the related gene numbers of the dominant genes in the BcaB and BcaC subgenomes. Orange indicates the number of terms and related genes in biological process; blue indicates those in molecular function; and green indicates those in cellular component.
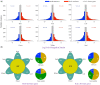
Figure 6
Analyses of NBS (nucleotide-binding site) family genes in Brassica carinata. A, Ks density plot of NBS family genes in Bca, Bra, Bni, Bol, Bju, Bna, and Arabidopsis (Ath). B, Connection lines of homologous NBS family gene pairs in the two diploid species B. nigra (BB) and B. oleracea (CC) and the tetraploid species B. carinata (BBCC). The orange lines represent the gene pairs with 0.1 < Ks < 0.2 and the green lines those with Ks ≤ 0.1. The left bar chart shows the numbers of NBS family genes in B. nigra, B. oleracea, and B. carinata; the right bar chart shows the numbers of pairs of NBS family genes with Ks < 0.2 in the six species in U’s triangle.
Figure 7
Whole-genome comparison of genes involved in GSL metabolic pathways in B. carinata and five other Brassica species in U’s triangle. A, Aliphatic and indolic GSL biosynthesis and catabolism pathways in B. carinata and the two other tetraploid species (B. napus, B. juncea). The copy numbers of GSL biosynthetic genes in B. carinata, B. juncea, and B. napus are listed in square brackets. Two important amino acid chain elongation loci MAM1/2 and ST5b with significantly different numbers are highlighted in U’s triangle models. B, Heat map of the log2-transformed number of GSL metabolic pathway-related genes in Arabidopsis, the six species in U’s triangle (blue font), and other Brassica species with genomes released. The range from yellow to blue represents a gradual increase in the number of genes. C–E, Maximum-likelihood trees of MAM, ST5, and AOP genes that were generated on the basis of amino acid sequences with 1,000 bootstrap repeats in Arabidopsis, B. carinata, and the five other species in U’s triangle.
Similar articles
- A chromosome-scale assembly of Brassica carinata (BBCC) accession HC20 containing resistance to multiple pathogens and an early generation assessment of introgressions into B. juncea (AABB).
Paritosh K, Rajarammohan S, Yadava SK, Sharma S, Verma R, Mathur S, Mukhopadhyay A, Gupta V, Pradhan AK, Kaur J, Pental D. Paritosh K, et al. Plant J. 2024 Jul;119(2):762-782. doi: 10.1111/tpj.16794. Epub 2024 May 9. Plant J. 2024. PMID: 38722594 - The final piece of the Triangle of U: Evolution of the tetraploid Brassica carinata genome.
Yim WC, Swain ML, Ma D, An H, Bird KA, Curdie DD, Wang S, Ham HD, Luzuriaga-Neira A, Kirkwood JS, Hur M, Solomon JKQ, Harper JF, Kosma DK, Alvarez-Ponce D, Cushman JC, Edger PP, Mason AS, Pires JC, Tang H, Zhang X. Yim WC, et al. Plant Cell. 2022 Oct 27;34(11):4143-4172. doi: 10.1093/plcell/koac249. Plant Cell. 2022. PMID: 35961044 Free PMC article. - Re-exploration of U's Triangle Brassica Species Based on Chloroplast Genomes and 45S nrDNA Sequences.
Kim CK, Seol YJ, Perumal S, Lee J, Waminal NE, Jayakodi M, Lee SC, Jin S, Choi BS, Yu Y, Ko HC, Choi JW, Ryu KY, Sohn SH, Parkin I, Yang TJ. Kim CK, et al. Sci Rep. 2018 May 9;8(1):7353. doi: 10.1038/s41598-018-25585-4. Sci Rep. 2018. PMID: 29743507 Free PMC article. - Insights on SNP types, detection methods and their utilization in Brassica species: Recent progress and future perspectives.
Yang S, Gill RA, Zaman QU, Ulhassan Z, Zhou W. Yang S, et al. J Biotechnol. 2020 Dec 20;324:11-20. doi: 10.1016/j.jbiotec.2020.09.018. Epub 2020 Sep 24. J Biotechnol. 2020. PMID: 32979432 Review. - Genome sequencing supports a multi-vertex model for Brassiceae species.
Cheng F, Liang J, Cai C, Cai X, Wu J, Wang X. Cheng F, et al. Curr Opin Plant Biol. 2017 Apr;36:79-87. doi: 10.1016/j.pbi.2017.01.006. Epub 2017 Feb 24. Curr Opin Plant Biol. 2017. PMID: 28242534 Review.
Cited by
- Molecular evolution analysis of MYB5 in Brassicaceae with specific focus on seed coat color of Brassica napus.
Dai G, Liu Y, Shen W, Zhu B, Chen L, Chen D, Tan C. Dai G, et al. BMC Plant Biol. 2024 Jan 16;24(1):52. doi: 10.1186/s12870-023-04718-6. BMC Plant Biol. 2024. PMID: 38229007 Free PMC article. - Improvement of glucosinolates by metabolic engineering in Brassica crops.
Miao H, Zeng W, Wang J, Zhang F, Sun B, Wang Q. Miao H, et al. aBIOTECH. 2021 Aug 7;2(3):314-329. doi: 10.1007/s42994-021-00057-y. eCollection 2021 Sep. aBIOTECH. 2021. PMID: 36303883 Free PMC article. Review. - Comprehensive genome-wide identification of Snf2 gene family and their expression profile under salt stress in six Brassica species of U's triangle model.
Qian F, Zuo D, Xue Y, Guan W, Ullah N, Zhu J, Cai G, Zhu B, Wu X. Qian F, et al. Planta. 2024 Jul 10;260(2):49. doi: 10.1007/s00425-024-04473-4. Planta. 2024. PMID: 38985323 - The evolution history of an allotetraploid mangrove tree analysed with a new tool Allo4D.
Wang Y, Li Y, Wu W, Shao S, Fang Q, Xu S, Guo Z, Shi S, He Z. Wang Y, et al. Plant Biotechnol J. 2024 Jun;22(6):1491-1503. doi: 10.1111/pbi.14281. Epub 2023 Dec 29. Plant Biotechnol J. 2024. PMID: 38157253 Free PMC article. - The Global Assessment of Oilseed Brassica Crop Species Yield, Yield Stability and the Underlying Genetics.
Zandberg JD, Fernandez CT, Danilevicz MF, Thomas WJW, Edwards D, Batley J. Zandberg JD, et al. Plants (Basel). 2022 Oct 17;11(20):2740. doi: 10.3390/plants11202740. Plants (Basel). 2022. PMID: 36297764 Free PMC article. Review.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
- Ban Y, Khan NA, Yu P (2017) Nutritional and metabolic characteristics of Brassica carinata Co-products from biofuel processing in dairy cows. J Agric Food Chem 65: 5994–6001 - PubMed
- Benderoth M, Pfalz M, Kroymann J (2008) Methylthioalkylmalate synthases: genetics, ecology and evolution. Phytochemistry Reviews 8: 255
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous