Long non‑coding RNA CASC15 facilitates esophageal squamous cell carcinoma tumorigenesis via decreasing SIM2 stability via FTO‑mediated demethylation - PubMed (original) (raw)
Long non‑coding RNA CASC15 facilitates esophageal squamous cell carcinoma tumorigenesis via decreasing SIM2 stability via FTO‑mediated demethylation
Bo Qin et al. Oncol Rep. 2021 Mar.
Abstract
Long non‑coding RNAs (lncRNAs) are involved in the regulation of esophageal squamous cell carcinoma (ESCC) progression. However, the function and mechanism of lncRNA cancer susceptibility candidate 15 (CASC15) are poorly defined. In the present study, tumor and normal adjacent tissues were collected from 45 patients with ESCC. Expression levels of CASC15, fat mass and obesity‑associated (FTO) protein and single‑minded 2 (SIM2) were examined via reverse transcription‑quantitative PCR and western blot assays. Cell proliferation and apoptosis were evaluated via MTT, flow cytometry and caspase‑3 activity assays, respectively. Additionally, an ESCC mouse xenograft model was used to assess the function of CASC15 in vivo. The interaction between FTO and CASC15/SIM2 was analyzed via RNA immunoprecipitation and RNA pull‑down assays. The results revealed that CASC15 expression was elevated in ESCC tissues, and patients with ESCC exhibiting high CASC15 expression had a poor prognosis. CASC15‑knockdown inhibited ESCC cell proliferation and facilitated apoptosis. Additionally, CASC15‑knockdown decreased the growth of ESCC xenograft tumors. CASC15 decreased SIM2 stability via FTO‑mediated demethylation. Additionally, FTO loss markedly weakened CASC15‑mediated pro‑proliferative and anti‑apoptotic effects in ESCC cells. SIM2 downregulation weakened the effect of CASC15‑knockdown on cell proliferation and inhibited the increase of the apoptotic rate and caspase‑3 activity induced by CASC15 depletion in ESCC cells. In conclusion, CASC15 promoted ESCC tumorigenesis by decreasing SIM2 stability via FTO‑mediated demethylation.
Keywords: esophageal squamous cell carcinoma; cancer susceptibility candidate 2; fat mass and obesity-associated protein; single-minded 2.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
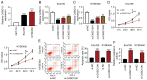
Figure 1.
CASC15 expression is increased in ESCC tumor tissues. (A) Expression analysis of CASC15 expression in ESCC was conducted using the Gene Expression Profiling Interactive Analysis database. (B) CASC15 expression was detected via reverse transcription-quantitative PCR in the tumor and adjacent normal tissues of patients with ESCC (n=45). (C) Overall survival of patients with ESCC was analyzed in high and low CASC15 expression groups. Data were expressed as the mean ± SD from three experiments. *P<0.05 and ***P<0.001. CASC15, cancer susceptibility candidate 15; ESCC, esophageal squamous cell carcinoma.
Figure 2.
Effects of CASC15-knockdown on ESCC cell proliferation and apoptosis. (A) CASC15 expression was determined via RT-qPCR in ESCC (Eca109 and KYSE450) and HET-1A cell lines. CASC15 expression was assessed via RT-qPCR in (B) Eca109 and (C) KYSE450 cells transfected with si-CASC15#1-3 or si-NC. Data analyzed via one-way ANOVA followed by Tukey's post-hoc test. (D) The proliferative ability of Eca109 and KYSE450 cells transfected with si-CASC15#1 or si-NC was assessed via an MTT assay at 0, 24, 48 and 72 h. Data analyzed via two-way ANOVA followed by Bonferroni's post-hoc test. (E) The apoptotic rate of Eca109 and KYSE450 cells transfected with si-CASC15#1 or si-NC was measured via flow cytometry. (F) Relative caspase-3 activity was examined in Eca109 and KYSE450 cells transfected with si-CASC15#1 or si-NC. Data were expressed as the mean ± SD from three experiments. **P<0.01 and ***P<0.001 vs. HET-1A or si-NC. CASC15, cancer susceptibility candidate 15; ESCC, esophageal squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; NC, negative control; OD, optical density.
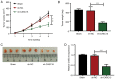
Figure 3.
Effect of CASC15-knockdown on esophageal squamous cell carcinoma xenograft tumor growth. (A) Tumor volume was detected every 7 days in each group. Data analyzed via mixed two-way ANOVA followed by Bonferroni's post-hoc test. (B) Tumor weight was measured on day 35 after injection in each group. (C) Representative images of tumors. (D) CASC15 expression was detected via reverse transcription-quantitative PCR in the tumor tissues of each group. Data analyzed via one-way ANOVA followed by Tukey's post-hoc test. ***P<0.001. CASC15, cancer susceptibility candidate 15; sh, short hairpin; NC, negative control.
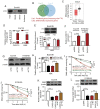
Figure 4.
CASC15 inhibited SIM2 expression and decreased SIM2 mRNA stability via FTO in esophageal squamous cell carcinoma cells. (A) RNA pull-down and western blotting assays were performed to determine the interaction of CASC15 and FTO protein. (B) Venn diagram of the possible mRNAs that interact with the FTO protein, as determined using the StarBase database and previous RNA-seq data. (C) SIM2 expression was assessed in ESCC tumor and adjacent normal tissues using the Gene Expression Profiling Interactive Analysis database. (D) CASC15 and SIM2 mRNA expression enriched by IgG or FTO antibodies were detected via RT-qPCR after a RIP assay in Eca109 cells. (E) Eca109 cells were transfected with pcDNA or the FTO overexpression vector. At 48 h post transfection, FTO protein expression was measured via western blotting, and m6A-enriched SIM2 and CASC15 mRNA expression was examined through RIP and RT-qPCR assays. Transfection efficiency of the (F) CASC15 overexpression vector and (G) si-FTO was measured in Eca109 and KYSE450 cell lines. (H) Eca109 cells were transfected with pcDNA, CASC15 overexpression vector, CASC15 overexpression vector + si-NC or CASC15 overexpression vector + si-FTO. At 48 h post transfection, RIP and RT-qPCR assays were conducted to measure SIM2 mRNA expression following m6A antibody enrichment. SIM2 mRNA stability was analyzed by performing an ActD assay in (I) Eca109 and (J) KYSE450 cells transfected with pcDNA, CASC15 overexpression vector, CASC15 overexpression vector + si-NC or CASC15 overexpression vector + si-FTO. Data analyzed via two-way ANOVA followed by Bonferroni's post-hoc test. SIM2 mRNA and protein expression was respectively examined via RT-qPCR and western blotting in (K) Eca109 and (L) KYSE450 cells transfected with pcDNA, CASC15 overexpression vector, CASC15 overexpression vector + si-NC or si-FTO. *P<0.05, **P<0.01 and ***P<0.001. CASC15, cancer susceptibility candidate 15; SIM2, single-minded 2; FTO, fat mass and obesity-associated; RIP, RNA immunoprecipitation; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; NC, negative control; m6A, n6-methyladenosine; ActD, Actinomycin D.
Figure 5.
Effect of FTO-knockdown on CASC15-mediated esophageal squamous cell carcinoma cell proliferation and apoptosis. Eca109 and KYSE450 cells were transfected with pcDNA, CASC15, CASC15+si-NC and CASC15+si-FTO. Subsequently, cell viability, apoptosis and caspase-3 activity were detected via (A and B) MTT (data analyzed via two-way ANOVA followed by Bonferroni's post-hoc test), (C and D) flow cytometry and (E) caspase-3 assays, respectively. *P<0.05, **P<0.01 and ***P<0.001. FTO, fat mass and obesity-associated; CASC15, cancer susceptibility candidate 15; si, small interfering RNA; NC, negative control; OD, optical density.
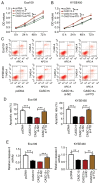
Figure 6.
CASC15 depletion decreased the proliferation and promoted the apoptosis of ESCC cells by upregulating SIM2. (A) SIM2 expression was measured via reverse transcription-quantitative PCR in tumor and adjacent normal tissues isolated from patients with ESCC (n=45). (B) Linear correlation of CASC15 and SIM2 expression in ESCC tissues. (C) SIM2 protein expression was detected in HET-1A, Eca109 and KYSE450 cells via western blotting. (D) SIM2 protein expression was measured by western blotting at 48 h post transfection in Eca109 and KYSE450 cells transfected with si-NC or si-SIM2. Eca109 and KYSE450 cells were transfected with si-NC, si-CASC15#1+si-NC or si-CASC15#1+si-SIM2. (E) SIM2 protein abundance was examined via western blotting. (F) Cell proliferation was determined by performing an MTT assay in Eca109 and KYSE450 cells. Data analyzed via two-way ANOVA followed by Bonferroni's post-hoc test. (G and H) Apoptotic rate was detected via flow cytometry. (I) Relative caspase-3 activity was detected using the caspase-3 assay kit in Eca109 and KYSE450 cells. *P<0.05, **P<0.01 and ***P<0.001. CASC15, cancer susceptibility candidate 15; ESCC, esophageal squamous cell carcinoma; SIM2, single-minded 2; si, small interfering RNA; NC, negative control; OD, optical density.
Similar articles
- RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma.
Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, Ma Y, Fang J, Wang Y, Cao W, Guan F. Cui Y, et al. J Exp Clin Cancer Res. 2021 Sep 20;40(1):294. doi: 10.1186/s13046-021-02096-1. J Exp Clin Cancer Res. 2021. PMID: 34544449 Free PMC article. - A novel long noncoding RNA, LOC440173, promotes the progression of esophageal squamous cell carcinoma by modulating the miR-30d-5p/HDAC9 axis and the epithelial-mesenchymal transition.
Wang G, Feng B, Niu Y, Wu J, Yang Y, Shen S, Guo Y, Liang J, Guo W, Dong Z. Wang G, et al. Mol Carcinog. 2020 Dec;59(12):1392-1408. doi: 10.1002/mc.23264. Epub 2020 Oct 20. Mol Carcinog. 2020. PMID: 33079409 - Roles of long non‑coding RNAs in esophageal cell squamous carcinoma (Review).
Yan Q, Wong W, Gong L, Yang J, Liang D, Chin KY, Dai S, Wang J. Yan Q, et al. Int J Mol Med. 2024 Aug;54(2):72. doi: 10.3892/ijmm.2024.5396. Epub 2024 Jul 4. Int J Mol Med. 2024. PMID: 38963019 Free PMC article. Review. - Single-cell RNA-sequencing data reveals the genetic source of extracellular vesicles in esophageal squamous cell carcinoma.
Zhou B, Guo W, Guo L, Li Y, Zheng Z, Huai Q, Tan F, Li Y, Xue Q, Ying J, Zhao L, Gao S, He J. Zhou B, et al. Pharmacol Res. 2023 Jun;192:106800. doi: 10.1016/j.phrs.2023.106800. Epub 2023 May 20. Pharmacol Res. 2023. PMID: 37217040 Review.
Cited by
- Long Non-coding RNAs With In Vitro and In Vivo Efficacy in Preclinical Models of Esophageal Squamous Cell Carcinoma Which Act by a Non-microRNA Sponging Mechanism.
Weidle UH, Birzele F. Weidle UH, et al. Cancer Genomics Proteomics. 2022 Jul-Aug;19(4):372-389. doi: 10.21873/cgp.20327. Cancer Genomics Proteomics. 2022. PMID: 35732324 Free PMC article. Review. - N6-methyladenosine Modification of Noncoding RNAs: Mechanisms and Clinical Applications in Cancer.
Ma M, Ye T, Wang J, Zhao H, Zhang S, Li P, Zhao G. Ma M, et al. Diagnostics (Basel). 2022 Nov 30;12(12):2996. doi: 10.3390/diagnostics12122996. Diagnostics (Basel). 2022. PMID: 36553003 Free PMC article. Review. - Crosstalk between lncRNAs and Wnt/β-catenin signaling pathways in lung cancers: From cancer progression to therapeutic response.
Wu T, Dong Y, Yang X, Mo L, You Y. Wu T, et al. Noncoding RNA Res. 2024 Mar 14;9(3):667-677. doi: 10.1016/j.ncrna.2024.02.013. eCollection 2024 Sep. Noncoding RNA Res. 2024. PMID: 38577016 Free PMC article. Review. - A study of the prognostic value of long non-coding RNA CASC15 in human solid tumors utilizing The Cancer Genome Atlas (TCGA) datasets and a meta-analysis.
Chen W, Qian W, Nie J, Dai M. Chen W, et al. Clin Exp Med. 2023 Feb;23(1):65-78. doi: 10.1007/s10238-021-00789-7. Epub 2022 Feb 1. Clin Exp Med. 2023. PMID: 35103883 - LncRNAs and Chromatin Modifications Pattern m6A Methylation at the Untranslated Regions of mRNAs.
Vaasjo LO. Vaasjo LO. Front Genet. 2022 Mar 17;13:866772. doi: 10.3389/fgene.2022.866772. eCollection 2022. Front Genet. 2022. PMID: 35368653 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
This study was supported by the National Natural Science Foundation of China (grant nos. 31801142 and 81900791), the Henan Key Project of Research and Development Plan (Science and Technology) (grant no. 192102310192) and the Henan Medical Science and Technology Joint Building Program (grant no. 2018020094).
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials